- Genetic structure and domestication of carrot (Daucus carota subsp. sativus) (Apiaceae). Origin in Central Asia, but no genetic bottleneck (sic).
- Data collection and assessment of commonly consumed foods and recipes in six geo-political zones in Nigeria: Important for the development of a National Food Composition Database and Dietary Assessment. Nigerians eat a lot of soup.
- The integration of crop rotation and tillage practices in the assessment of ecosystem services provision at the regional scale. Good trick if you can do it.
- Nutritional composition of minor indigenous fruits: Cheapest nutritional source for the rural people of Bangladesh. If only the rural people knew about this.
- Effectiveness of selection at CIMMYT’s main maize breeding sites in Mexico for performance at sites in Africa and vice versa. Is high. Phew.
- Olive trees as bio-indicators of climate evolution in the Mediterranean Basin. Olives in Germany by 2100?
- Crop genetic diversity benefits farmland biodiversity in cultivated fields. Mixed wheat fields better for soil invertebrate biodiversity than fields with single varieties.
- IT background of the medium-term storage of Martonvásár Cereal Genebank resources in phytotron cold rooms. The interesting thing is that the system links genebank data with breeders’ data. Don’t see that a lot.
Conserving Prunus africana?
I’ve been sitting on it for a while, but a paper which AoB Blog discussed back in January led me to uncover a whole load of stuff on Prunus africana. The African Cherry Tree does not rate a leaflet in the African Food Tree Species series, perhaps because it’s not a, well, food tree, but that doesn’t mean it’s not important.
Chemicals extracted from the tree’s bark are used in a range of pharmaceutical products to treat enlarged prostate (benign prostatic hyperplasia), an extremely common condition that affects up to half of men aged over 50.
Hence various efforts to develop sustainable harvesting methods. And also an interesting series of diversity and demographic studies:
- Phylogeography of the Afromontane Prunus africana reveals a former migration corridor between East and West African highlands: “The high genetic similarity found between western Uganda and west African populations indicates that a former Afromontane migration corridor may have existed through Equatorial Africa.”
- Structural diversity and regeneration of the endangered Prunus africana (Rosaceae) in Zimbabwe: “…poor regeneration, fewer P. africana trees in small and large size classes, dominance of positive height and diameter differentiation and high mingling.”
- Divergent pattern of nuclear genetic diversity across the range of the Afromontane Prunus africana mirrors variable climate of African highlands: “The observed patterns indicate divergent population history across the continent most likely associated to Pleistocene changes in climatic conditions. The high genetic similarity between populations of West Africa with population of East Africa west of the Eastern Rift Valley … provides further evidence for a historical migration route. Contrasting estimates of recent and historical gene flow indicate a shift of the main barrier to gene flow from the Lake Victoria basin to the Eastern Rift Valley…”
- Modelling the potential distribution of endangered Prunus africana (Hook.f.) Kalkm. in East Africa: “Prunus africana distribution is thus highly vulnerable to a warming climate and highlights the fact that both in-situ and ex-situ conservation will be a solution to global warming.”
Maybe we could do with some more seed behaviour data. But it would seem there is now plenty of diversity, demographic and sustainable harvesting information on which to base a comprehensive conservation strategy. Is someone coming up with one?
Wheat diversity collections seen and unseen
A couple of things on wheat today, thanks to Tom Payne at CIMMYT, our go-to guy for all things triticaceous. First, Kew’s new page on Bread Wheat, which has a lot of useful information, including this:
About 250,000 samples of bread wheat are held in agricultural gene banks around the world, so the plant is far from being threatened. However, there is cause for concern in terms of bread wheat landraces, which are being replaced by modern cultivars and under threat of extinction if not already conserved in ex-situ collections.
The figure is I suspect from Genesys, from which the map below is taken. WIEWS, which covers many more genebanks, gives 546,797, but there’s probably much more duplication in that number than in the Genesys one.
And second, from the just published study “Agricultural Innovation: The United States in a Changing Global Reality,” a wide-ranging analysis of the benefits to the US of investment in international agricultural research, a discussion of the pedigree of the hard red winter wheat variety Jagger, the most widely planted wheat variety in the United States:
The breeders who developed Jagger drew on genetic material from all over the world and throughout the United States. Jagger was formed by crossing the breeding line KS82W418 (developed by the Kansas agricultural experimental station) with the variety Stephens (developed jointly by the Oregon agricultural experiment station and USDA-ARS). In turn, these two varieties stand firmly on the shoulders of the investments in scientific crop breeding over the past century and the eons of selection and seed-saving efforts of farmers since wheat was first domesticated around 10,000 years ago. Jagger’s ancestry includes varieties like Turkey Red from Russia, Noe from France, Federation and Purplestraw from Australia, Yaqui from Mexico, and Etawah from India.
Too bad that the closest the authors come to saying where those ancestors of Jagger, along with their 250,000 or 500,000 or whatever cousins, may be found, despite numerous references in the text to CIMMYT and USDA, is this laconic sentence:
In addition to the efforts of private citizens, the US Department of Agriculture (USDA) sent its scientists to the far corners of the globe in search of better plant varieties.
Maybe I’ll send them that Kew link.
US crop wild relatives inventoried
Our friend Colin Khoury and his associates have a paper out in Crop Science on inventorying crop wild relatives in the US. The press release that goes with it is getting picked up. The bottom line is easy to summarize, and Colin does so in the abstract:
We prioritize 821 taxa from 69 genera primarily related to major food crops, particularly the approximately 285 native taxa from 30 genera that are most closely related to such crops. Both the urgent collection for ex situ conservation and the management of such taxa in protected areas are warranted, necessitating partnerships between concerned organizations, aligned with regional and global initiatives to conserve and provide access to CWR diversity.
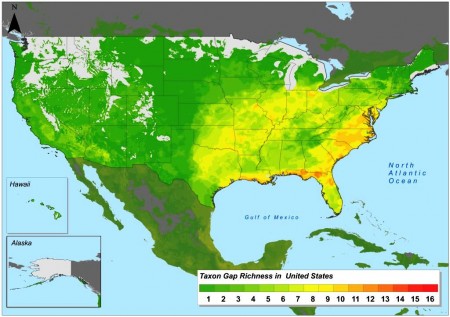
But where to start with all that collecting and in situ work? Well, here’s a little peek at the next phase of Colin’s work, which will answer that question. He’ll be modelling the distribution of the priority species using the GIS resources at CIAT, and mapping areas of high species diversity, and also areas which are under-represented in conservation efforts (gaps). Using just a portion of the data, and therefore yielding only very preliminary results, this is the sort of thing that comes out:
We look forward to the final results in due course. Good luck, Colin, and thanks for the sneak preview.
Tracing the Polynesian migrations through DNA, but not only
I know you probably don’t have an hour to spare to listen to a lecture on the evidence for pre-Columbian contacts between Polynesians and South American cultures, but Dr Lisa Matisoo-Smith does a really good job of galloping though the DNA and archaeological evidence from humans, commensals and livestock in a recent podcast from the Bishop Museum. She even mentions crops.
The bottom line? The human anatomical and artifact evidence is compelling, but the DNA is not cooperating yet. At least the human DNA. But listen to it. While you’re preparing dinner or something. I just wish the Bishop had thought to put the slides online too.