The Global Panel on Agriculture, Food Systems and Nutrition has come out with a Statement on Climate Change, Food Systems and Nutrition, in the run-up to a side event at COP21 in Paris. Nothing particularly surprising there, but I just wanted to highlight two things. First, I was impressed by how the problem was set out:
Climate change is expected to push down global farm output by 2% per decade between now and 2050. Demand for food is expected to rise substantially during that same period.
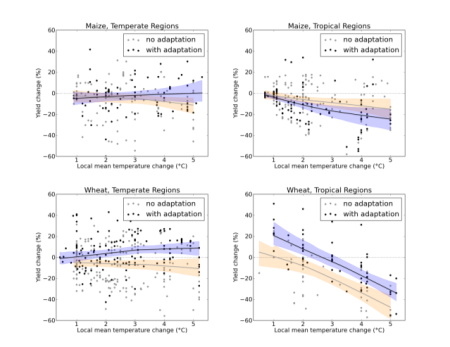
Yes, those fancy IPCC synthesis diagrams (see below) are cool and everything, but there’s a lot to be said for the directness and starkness of those two sentences. Sometimes a few well-chosen words are better than a picture.
Then there are the policy recommendations:
1. Include diet quality goals with adaptation targets proposed for climate action.
2. Diversify agricultural investments, factoring in the local realities of ecological sustainability and comparative advantage.
3. Support greater food system efficiency so that outputs per unit of water, energy, land, and other inputs are optimised and the footprint of agriculture and non-farm activities are better managed to meet both food demand and higher-quality diets.
4. Integrate measures to improve climate change resilience and the nutritional value of crop and livestock products along the value chain, from production to marketing.
5. Protect the diet quality of the poor in the face of supply shocks and growing food demand through social protection, for example.
6. Promote the generation and use of rigorous evidence on appropriate investments along food value-chains, which are resilient to climate change and also deliver positive dietary outcomes and support improved nutrition.
And it’s striking how agricultural biodiversity in one way or another could be argued to underpin all of these.
