- Getting to grips with citrus greening.
- A review of the effect of oil palm, soybean, and jatropha production on biodiversity and ecosystem functions in tropical forests is in the offing. Here’s what will be done.
- Sweet potatoes are good for the health of children. If you can get them to eat the things.
- The history of monogerm sugar beet.
- There’s no best way to link poverty alleviation and biodiversity conservation. The Global Meeting of the Indigenous Peoples’ Forum came up with a few ways, I’m sure.
- Vegetarian tribe in India is certainly doing that conservation, but the poverty alleviation?
- Jeremy takes a stroll around the allotment.
- Musalit is back in the game. And speaking of which, who would not have wanted to be at the Makira Banana Festival?
- Squeezing the most out of Russet Burbank. Oh, look at some new landraces, already!
- Portugal becomes second genebank to adopt GRIN-Global.
- AVRDC is looking for its next leader. Must know his/her onions.
Peachy photos online
Not only is U.P. Hedrick’s magisterial The Peaches of New York on Open Library. The images are also on Flickr, under Internet Archive Book Images. Mouthwatering.
Brainfood: Grape genetics & conservation, Ecosystem restoration & services, Collecting cycads, Pigeonpea genomics, Wild pigeonpea gaps, Breadfruit collection diversity, Banana collection diversity, Conserving mammals, Bhutanese cereal diversity, Potato nutrition
- The preservation of genetic resources of the vine requires cohabitation between institutional clonal selection, mass selection and private clonal selection. Intra-varietal diversity, that is. They apparently do it best in Portugal.
- Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Cool things I learned from this paper: 1. Pinot Meunier is a periclinal mutant. 2. Tendrils are inflorescences. 3. Dwarf grapes are dwarf for the same reason dwarf wheat is dwarf. No word on what the Portuguese are doing about it. And the fact that the paper is over 10 years old is irrelevant to its coolness.
- Quantifying the impacts of ecological restoration on biodiversity and ecosystem services in agroecosystems: A global meta-analysis. Supporting ecosystem services go up 40% on average, regulating ES by 120%. Well worth having.
- Can a Botanic Garden Cycad Collection Capture the Genetic Diversity in a Wild Population? Yes, but need to “(1) use the species biology to inform the collecting strategy; (2) manage each population separately; (3) collect and maintain multiple accessions; and (4) collect over multiple years.” Maybe they should talk to the Xishuangbanna Tropical Botanical Garden guys.
- Genebanking Seeds from Natural Populations. It’s more difficult than with crops. And then you’ve got the above.
- Genomics-assisted breeding for boosting crop improvement in pigeonpea (Cajanus cajan). “…pigeonpea has become a genomic resources-rich crop and efforts have already been initiated to integrate these resources in pigeonpea breeding.” And now we wait.
- Crop wild relatives of pigeonpea [Cajanus cajan (L.) Millsp.]: Distributions, ex situ conservation status, and potential genetic resources for abiotic stress tolerance. 15 of them need collecting. And, presumably, genotyping (see above).
- Diversity in the breadfruit complex (Artocarpus, Moraceae): genetic characterization of critical germplasm. 349 individuals from 255 accessions are 197 unique genotypes from 129 lineages.
- Musa spp. Germplasm Management: Microsatellite Fingerprinting of USDA–ARS National Plant Germplasm System Collection. Used to identify mislabelled in vitro accessions. Data in GRIN-Global.
- Education and access to fish but not economic development predict chimpanzee and mammal occurrence in West Africa. Having fish to eat means you leave chimpanzees alone.
- Community Perspectives on the On-Farm Diversity of Six Major Cereals and Climate Change in Bhutan. About 30% of varieties lost over last 20 years. But what does it mean that “93% of the respondents manage and use agro-biodiversity for household food security and livelihood”? What exactly does that other 7% of farmers do?
- Amino acid composition and nutritional value of four cultivated South American potato species. S. goniocalyx is best for you. But does it taste any good?
Nibbles: Old breweries, Old grape seeds, New beer, Sheep breeds, Indian rice landraces, GM rice in China, Barley breeding, Botanical tipple, Mata Atlantica conservation, Quinoa
- There are some really old breweries out there.
- And some really old grape seeds in Sardinia.
- The beauty of hops. New and old.
- Some very photogenic old sheep breeds in the Lake District.
- Someone has discovered some old salt-tolerant rice landraces. Also medicinal and aromatic rices. Well I never.
- Meanwhile, at the other end of the rainbow, the Chinese dip a toe into gourmet GM rice.
- Brit boffins breed flood-proof barley.
- Botanical gin. Two of my favourite things, combined.
- Saving the Pau Brasil is, well, complicated. But what else was it going to be?
- Yet another roundup of the pros and cons of quinoa.
New version of banana genebank information system goes live
Great to see a new version of the Musa Germplasm Information System (MGIS) released. The URL is unchanged. The key improvements are listed as follows (slightly edited):
1. All information on a single accession can be viewed in one page
2. Taxonomic content of each collection is summarized graphically.
3. Easier data filtering and export functions.
4. Users can share comments on any accession.
5. Accessions can be requested online via the Musa Online Requesting system (MORS) with a modified interface.
I particularly like the ability to comment, though you do have to register for that. The data cover 2,281 accessions from six genebanks around the world, 1 including 1,456 in the International Transit Centre (ITC) managed by Bioversity International in Belgium:
The ITC data are also in Genesys, which shows 1,529 accessions rather than 1,456. I assume MGIS is the more up to date, but I’m unclear why there should be a difference. 2
You can search among the 2,281 accessions on name or number; or by filtering by any combination of genebank, species, subspecies, genome group (AAB, say), subgroup (Cavendish, say), country of origin, ploidy, whether there’s a photo, whether it’s been included in a molecular study, and availability. Searching is pretty fast.
Each accession gets a nice page summarizing all the pertinent information.
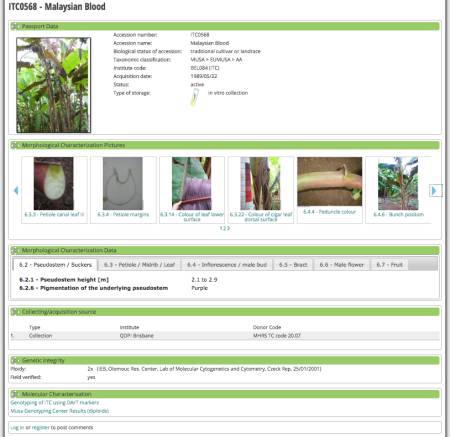
That information can include morphological characterization data, and illustrations, as you can see above, but I could not find a way of searching the database based on a particular descriptor or combination of descriptors. You get a map when collecting locality is known, but you can’t map multiple accessions, as far as I could see. You’d have to do that in Genesys, I guess.
If you want to download data, you have to cut and paste accession numbers into a form on another page, and then you get a CSV or XLS. It didn’t look to me like you could export either morphological characterization data or molecular data. I have to say I was disappointed by the whole export thing.
So, some good things, some not so good things in this new version of MGIS. I’ll be keeping an eye on it for further developments. And continue playing with it, of course. Maybe I missed something.

