- UN Special Rapporteur on food thinks “questions of the 60s are not the questions of today.” Does he think the CGIAR is answering the questions of the 60s? One suspects so, but surely there are points of agreement, e.g. nutrition, food systems, natural resources management…
- Farmers would be willing to pay quite a premium for drought tolerant (DT) rice hybrids, but for DT varieties not so much. That’s an opportunity for public-private partnerships. Or is that a 60s answer to a 60s question?
- Mr de Schutter probably knows all about this bibliography of agroecology in action. Which all seems so much more 60s than hybrid rice somehow.
- How 60s is it to want to produce decent amaranthus seed? It’s totally unfair, but I can’t resist linking to this now.
- Ethiopian genebank, set up in response to the genetic erosion of the 60s, gets nice, long writeup in The Guardian by way of introduction to a bare-bones couple of final paragraphs on some G8 poverty reduction plan. Nice video though.
- There was no Facebook in the 60s for genebanks to strut their stuff on.
CGN18108 it is
Simon Foster very kindly took the trouble to post a comment setting the record straight on the source of that blight gene:
Apologies, a previous tweet from ourselves erroneously confirmed the accession as CGN18000. It is in fact CGN18108 which is still listed in the database as Solanum okadae (was subsequently found to be S. venturii in DNA fingerprinting studies).
The origin of Rpi-vnt1 is detailed in the original research paper describing the cloning and characterisation of the gene and which is cited in the Roy. Soc. paper published yesterday. All acknowledgement of sources was published in that paper.
Foster SJ, Park T-H, Pel M, Brigneti G, Sliwka J, Jagger L, van der Vossen E, Jones JDG. 2009 Rpi- vnt1.1, a Tm-2(2) homolog from Solanum venturii, confers resistance to potato late blight. MPMI 22, 589 – 600. (doi:10.1094/MPMI-22-5-0589)
Here’s the relevant bit of that paper:
Accessions of S. venturii and S. okadae were obtained from the Centre for Genetics Resources in Wageningen, the Netherlands (CGN) (Table 1). The S. venturii accessions were originally listed as S. okadae in the CGN database but have recently been reclassified based on work using amplified fragment length polymorphism (AFLP) markers to study the validity of species labels in Solanum section Petota (Jacobs 2008; Jacobs et al. 2008).
So my apologies to Dr Foster. There is indeed a very full and proper acknowledgement of the source of the gene in the earlier paper. However, I do still think that it would not really have taken much effort to also include an acknowledgement in the later paper. The confusion over which accession was actually used that I fell into, admittedly without taking the trouble of following the references, is evidence of why it’s important to do so.
Now to suggest to CGN that they may want to change the species name of CGN18108 in their database…
LATER: Just realized we started talking about all this quite a while ago.
Credit where credit is due
![]() You may have been following the reaction to the media frenzy about the announcement yesterday of a new way of making potatoes resistant to blight, cause of the Irish Famine a century and a half ago. I won’t add to the confusion, beyond saying that, as in everything in life, there are always alternatives, and wouldn’t it be nice if we gave them all a fair go?
You may have been following the reaction to the media frenzy about the announcement yesterday of a new way of making potatoes resistant to blight, cause of the Irish Famine a century and a half ago. I won’t add to the confusion, beyond saying that, as in everything in life, there are always alternatives, and wouldn’t it be nice if we gave them all a fair go?
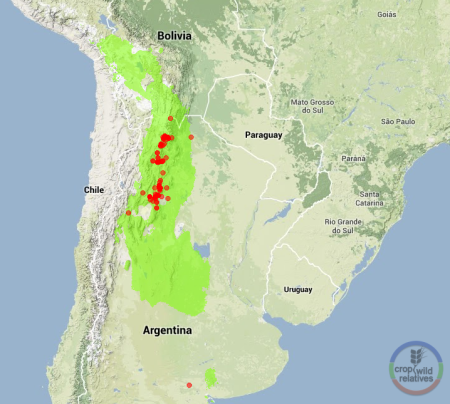
No, what I want to talk about here is where the gene in question came from, the gene for blight resistance that was inserted into the popular British variety Desiree to such apparently good effect. We know from the paper that the gene is called Rpi-vnt1.1 and that it came from the potato wild relative Solanum venturii, but that’s all. I think that’s insufficient. There may be a reference in the paper or the supplementary material 1 to the source of the Solanum venturii germplasm that was used in this work, but I was unable to find it. Apologies to the authors if I missed it, and thanks to whoever it was at the Sainsbury Lab that tweeted a response to my request for information:
@AgroBioDiverse @Cipotato Seed was from: http://t.co/XLC5Ud82mh
— The Sainsbury Laboratory (@TheSainsburyLab) February 17, 2014
It turns out that the germplasm was obtained from the Centre for Genetic Resources, the Netherlands, which has often been mentioned on this blog. Most recently, as coincidence would have it, yesterday, and on the subject of potatoes to boot. CGN is at the very forefront of genebank documentation, so it was very easy to find that there are 3 accessions of the species in question at CGN, that only one of those is resistant to blight, that it is CGN18000, that it was collected in 1972 in Argentina at an elevation of 3200m and that it came to CGN via the Institut fur Pflanzenbau und Pflanzenzuchtung Bundesforschungsanstalt, Braunschweig, Germany. There are more accessions out there (although apparently not at CIP, the International Potato Center, at least according to Genesys), and also herbarium specimens. Enough in fact to come up with an estimate of the overall geographic distribution of S. venturii, and perhaps identify some gaps.
Or am I wrong? It’s an informed guess. If there had been more than one accession with resistance, it would have been difficult to know which one was actually used in the paper. Anyway, I look forward to the day when even the authors of whizz-bang biotech papers will spend five minutes to do what I just did and properly acknowledge, perhaps even link to, the genebank, and the specific accession, whose existence ultimately allowed them to do their work.
Brainfood: Value of Chiloé, Zimbabwe sorghum, Rosa karyotypes, PSM diversity, Pear diversity, Medic clines, Wild rices, Barley adaptation, Coffee agroforesty
- Valuing cultural ecosystem services: Agricultural heritage in Chiloé island, southern Chile. Willingness to pay at US$50.5 per person per year, and not related to distance from site.
- Assessments of genetic diversity and anthracnose disease response among Zimbabwe sorghum germplasm. New sources of resistance (for the US) in even a moderately diverse collection.
- Karyotype Analysis of Wild Rosa Species in Xinjiang, Northwestern China. It’s just amazing to me that people still do karyopypes.
- Explaining intraspecific diversity in plant secondary metabolites in an ecological context. Trait variance in these things is considerable, partly genetic and can evolve, maybe even faster than mean trait values.
- Identifying genetic diversity and a preliminary core collection of Pyrus pyrifolia cultivars by a genome-wide set of SSR markers. Close relationship between China and Japan, and Sichuan a bit of a nexus.
- Genomic Signature of Adaptation to Climate in Medicago truncatula. Found genes associated with position along 3 environmental clines in a set of populations, then were able to predict performance of other populations based on genotype.
- Could abiotic stress tolerance in wild relatives of rice be used to improve Oryza sativa? Yes, and from these particular places.
- An efficient method of developing synthetic allopolyploid rice (Oryza spp.). Should make using those wild relatives a bit easier.
- Can barley (Hordeum vulgare L. s.l.) adapt to fast climate changes? A controlled selection experiment. Maybe not. Not even the landrace.
- Coffee landscapes as refugia for native woody biodiversity as forest loss continues in southwest Ethiopia. “Coffee farms could support a considerable portion, though not all, of the woody biodiversity of disappearing forests.” No word on what it does to the coffee, though.
Nibbles: New potatoes, Wild species, Native maize, Conservation course, Indigenous fishery, Yield trends
- Wild relative rescues potatoes. Which wild relative? Well for that you’ll have to read the paper. The FAQ on that. Or if you want an alternative. More the better, I guess. And just to remember what makes it all possible: diversity in fields and genebanks.
- Wild species not just useful to food security as sources of genes, of course. And more.
- Indigenous peoples save corn.
- Maybe some of them would be interested in this MSc at Bangor.
- Indigenous peoples can catch — and save? — fish after all.
- So is there stagnation in yield increases or what? Lobell reviews book that says maybe not.