- The industrial sliced loaf as racist fantasy.
- Bill Gates talks dirty. About cassava, settle down.
- Where the wild things are is where the languages are, but why? And where are they endangered?
- This new Indian wild banana is probably a bit endangered.
- The past may be a foreign country, but they got Street View there too.
- Blockbuster rice in India. (But how energy-efficient is it?)
- And potentially blockbuster livestock breeds in Kenya.
- China goes GMO. Which of these?
- Maybe they should read this vision for organic farming? You know, just for completeness?
- Have a food adventure! Just perhaps not in China.
- FAO and National Geographic have a food security adventure together. For more stuff like this, no doubt…
- Eat more plants, and ditch the junk food. Sounds easy, doesn’t it? Ok, for the more complicated, nerdy approach, there’s always fancy indicators.
- Ethiopia’s community seed banks.
- I bet there are some in Zimbabwe…
- African Development Bank makes a bet on agroforestry. Maybe health is why? There’s more that one reason…
- Gotta be strategic with your legume collecting.
- Want conservation science to translate into impact? Don’t publish behind a paywall.
- CIMMYT earns its keep.
- Build a better apple, and you won’t be able to keep the journalists away.
Comparing the niches of wild, feral and cultivated tetraploid cottons, Conclusion
We’re trying something new for us this week. Dr Geo Coppens co-authored an interesting paper 1 recently which brings together a number of our concerns: domestication, diversity, crop wild relatives, spatial analysis… He’s written quite a long piece about his research, which we’ll publish here in three instalments. This is the third and final instalment. Here’s the previous one.
Understanding the causes of the large difference between the distributions of TWC and that of cultivated and feral cottons is crucial for future breeding efforts. On one hand, the strong genetic divergence between wild and feral G. hirsutum that we have found in a sister SSR marker study may be partly related to differential selection and adaptation, and/or to the difficulty for domesticated forms to get established in a hostile environment. On the other hand, there is little doubt that much of the difference lies in the distinct ecological interpretation to be given to their respective models. Indeed, we may consider that the distribution of cultivated cottons approaches that of the fundamental niche of the species (i.e. where abiotic factors are favourable), which is obviously wider than the realized niche (further constrained by biotic factors, including competition).
Indeed, biotic factors are much less important under cultivation. For feral plants, we may refer to another niche concept: source-sink relations between cultivated and feral populations may explain how the latter may be found in unsuitable habitats (Pulliam, 2000). Logically, escapes from cultivation simply persist in the vicinity of cultivated cottons, in recently abandoned fields or along roadsides, benefitting from the open agricultural landscape. Why do feral populations observed further from cotton fields and dooryards, in more developed secondary vegetation (our ‘wild/feral’ category), present a very similar climatic envelope, and not an intermediate one? In fact, once interspecific competition has been avoided or reduced at the seedling stage, ancient escapes may persist for long, even in relatively high secondary vegetation. This is consistent with germplasm collectors’ reports (e.g. Ulloa et al., 2006) that feral populations are morphologically similar to local races of cultivated cotton, and that they are getting rarer as the cultivation of perennial cotton declines in Mexico.
The cotton example shows the relevance of ecological niche concepts even in the seemingly straightforward study of crop plant distribution, and particularly in the context of domestication studies, where it is crucial to distinguish between wild and feral forms. Comparing cultivation areas of domesticated forms with the original distribution of their wild relatives does not allow direct inferences on crop potential for climatic adaptation. Getting back to the tetraploid cotton example, niche conservatism after more than 1 million years of species diversification (the age of the AADD allotetraploid Gossypium genome) should make us more cautious in evaluating the result of 10,000 years of breeding on crop climatic adaptation. While there is no doubt that domestication and cultivation have resulted in widening of crops’ climatic envelopes, we need to understand much better the respective shares of genome and environment in the process. In the face of global climate change, we will need the tools of both genetic improvement and agroecology.
References
Brubaker CL, Wendel JF (1994) Reevaluating the origin of domesticated cotton (Gossypium hirsutum, Malvaceae) using nuclear restriction fragment length polymorphism (RFLP). Am J Bot 81: 1309–1326. doi: 10.2307/2445407
Coppens d’Eeckenbrugge G, Lacape J-M (2014) Distribution and Differentiation of Wild, Feral, and Cultivated Populations of Perennial Upland Cotton (Gossypium hirsutum L.) in Mesoamerica and the Caribbean. PLoS ONE 9(9): e107458. doi:10.1371/journal.pone.0107458
Fryxell PA (1979) The Natural History of the Cotton Tribe (Malvaceae, Tribe Gossypieae): Texas A&M University Press, College Station, TX. 245 p.
Hutchinson JB (1954) New evidence on the origin of the old world cottons. Heredity 8: 225–241. doi: 10.1038/hdy.1954.20
Hutchinson JB, Silow RA, Stephens SG (1947) The evolution of Gossypium and the differenciation of the cultivated cottons: Emprire Cotton Growing Corporation. 160 p.
Miller AJ, Knouft JH (2006) GIS-based characterization of the geographic distributions of wild and cultivated populations of the Mesoamerican fruit tree Spondias purpurea (Anacardiaceae). Am J Bot 93: 1757–1767. doi: 10.3732/ajb.93.12.1757
Pulliam HR (2000) On the relationship between niche and distribution. Ecology Letters 3: 349–361. doi: 10.1046/j.1461-0248.2000.00143.x
Russell J, van Zonneveld M, Dawson IK, Booth A, Waugh R, et al. (2014) Genetic Diversity and Ecological Niche Modelling of Wild Barley: Refugia, Large-Scale Post-LGM Range Expansion and Limited Mid-Future Climate Threats? PLoS ONE 9(2): e86021. doi:10.1371/journal.pone.0086021
Sauer JD (1967) Geographic reconnaissance of seashore vegetation along the Mexican Gulf coast; Mc Intire WG, editor. Baton Rouge. 59 p.
Stephens SG (1965) The effects of domestication on certain seed and fiber properties of perennial forms of cotton, Gossypium hirsutum L. The American Naturalist 94: 365–372. doi: 10.1086/282377
Thomas E, van Zonneveld M, Loo J, Hodgkin T, Galluzzi G, et al. (2012) Present Spatial Diversity Patterns of Theobroma cacao L. in the Neotropics Reflect Genetic Differentiation in Pleistocene Refugia Followed by Human-Influenced Dispersal. PLoS ONE 7(10): e47676. doi:10.1371/journal.pone.0047676
Ulloa M, Stewart JM, Garcia EA, Godoy S, Gaytan A, et al. (2006) Cotton genetic resources in the Western states of Mexico: in situ conservation status and germplasm collection for ex situ preservation. Genet Resour Crop Evol 53: 653–668. doi: 10.1007/s10722-004-2988-0
Wendel JF, Brubaker CL, Seelenan T (2010) The origin and evolution of Gossypium. In: Stewart JM, Oosterhuis DM, Heitholt JJ, Mauney JR, editors. Physiology of cotton. Dordrecht: Spinger, Netherlands. pp. 1–18.
Brainfood: Shea diversity, Weedy rice, Old barley, Grassland diversity, Afrikaner cattle, Sudanese baobab, FGR review, Small farmers, Ecological pest management, Andean ag changes, European soil biodiversity, Durum adaptation, Agrobodiversity indicators
- Genetic diversity in shea tree (Vitellaria paradoxa subspecies nilotica) ethno-varieties in Uganda assessed with microsatellite markers. Three geographic populations revealed by SSRs, not much related to the folk classification.
- Malaysian weedy rice shows its true stripes: wild Oryza and elite rice cultivars shape agricultural weed evolution in Southeast Asia. The weed is caught in the middle and swings both ways.
- Farmers without borders—genetic structuring in century old barley (Hordeum vulgare). Nordic barley structured latitudinally, with lots more variation within landrace accessions than now, according to historical specimens.
- The Agrodiversity Experiment: three years of data from a multisite study in intensively managed grasslands. Does increasing plant diversity in intensively managed grassland communities increase their resource use efficiency? No idea, but here’s the data from a huge multi-site experiment. Go crazy.
- Genetic diversity in selected stud and commercial herds of the Afrikaner cattle breed. It’s doing just fine, genetically, despite recent declines in numbers.
- The African baobab (Adansonia digitata, Malvaceae): Genetic resources in neglected populations of the Nuba Mountains, Sudan. Maybe a little more variation in homesteads compared to the wild. Maybe.
- Seeing the trees as well as the forest: The importance of managing forest genetic resources. The first State of the World’s Forest Genetic Resources and the first Global Plan of Action for the Conservation, Sustainable Use and Development of Forest Genetic Resources summarized: exchange, test in common gardens, and be clever with genetics, in breeding, management and restoration.
- Are small family farms a societal luxury good in wealthy countries? Rich countries don’t mind inefficient farms because they look nice.
- DIVECOSYS: Bringing together researchers to design ecologically-based pest management for small-scale farming systems in West Africa. Where do I sign up?
- Ecosystem governance in a highland village in Peru: Facing the challenges of globalization and climate change. Big Dairy doing for Andean crops.
- Intensive agriculture reduces soil biodiversity across Europe. What they said.
- The climate of the zone of origin of Mediterranean durum wheat (Triticum durum Desf.) landraces affects their agronomic performance. 4 main climatic zones, accounting for up to 30% of variation in important evaluation traits. FIGS, anyone?
- Indicators for the on-farm assessment of crop cultivar and livestock breed diversity: a survey-based participatory approach. And only 5 of them too!
Speaking about Speaking of Food
This special issue, “Speaking of Food: Connecting basic and applied plant science,” aims to provide concrete examples of how a wide range of basic plant science, the types of scientific studies commonly published in AJB, are relevant for the future of food. This Special Issue was inspired by Elizabeth A. Kellogg’s 2012 Presidential Address to the Botanical Society of America, and resulted in part from a symposium and colloquium by the same name that took place at the 2013 Botany meetings in New Orleans, LA. The issue editors are grateful to the Botanical Society of America, the American Society of Plant Taxonomists, and the Torrey Botanical Society for support of this work.
Special issue of the American Journal of Botany, that is. Alas, only one of the papers, the one on strawberries, is open access, though.
Comparing the niches of wild, feral and cultivated tetraploid cottons, Part 2
We’re trying something new for us this week. Dr Geo Coppens co-authored an interesting paper 1 recently which brings together a number of our concerns: domestication, diversity, crop wild relatives, spatial analysis… He’s written quite a long piece about his research, which we’ll publish here in three instalments. This is the second instalment. Here’s the first, in case you missed it.
To investigate the origin of cultivated cotton in the New World, we also had to define the distribution of the wild precursors, which meant distinguishing True Wild Cotton (TWC) populations from feral (secondarily wild) populations. We first defined five categories: (1) TWC populations, which were described as such by cotton experts with consistent ecological and/or morphological criteria; (2) wild/feral cottons, reported to grow in protected areas and/or forming relatively dense populations in secondary vegetation; (3) feral plants living in anthropogenic habitats (field and road margins); (4) cultivated perennial cottons; and (5) plants of indeterminate status. We then organized our extensive dataset of locality information from herbaria and genebanks in the same way as Russian nesting dolls.
Avoiding the problematic comparisons between consecutive categories, we first modelled G. hirsutum distribution based on the whole sample, then reducing the sample to ‘feral’+‘wild/feral’ + TWC , then to ‘wild/feral’+TWC, and finally to TWC alone. Thus, we could see that the distribution model (we used the Maxent software for niche modelling) was not significantly affected by the successive removals of specimens classified as indeterminate, cultivated, or feral. Only the model based on TWC populations diverged very clearly, their distribution being severely restricted to a few coastal habitats, in northern Yucatán and in the Caribbean, from Venezuela to Florida (Figures 1 and 2).
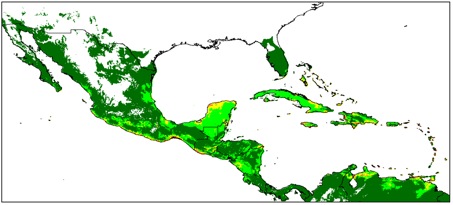
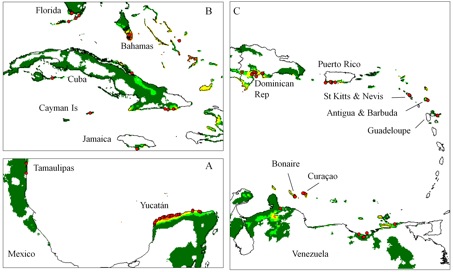
A principal component analysis on climatic parameters confirmed the absence of clear differences among the climatic niches of cultivated, feral, and ‘wild/feral’ populations, contrasting strikingly with that of TWC populations, restricted to the most arid coastal environments. The TWC habitat’s harsh conditions are quite certainly related to the fact that “Gossypium is xerophytic and that even its most mesophytic members are intolerant of competition, particularly in the seedling stage” (Hutchinson, 1954). The proximity to the sea plays a role itself, for these “extreme outpost plants” (Sauer, 1967). Indeed, as Fryxell (1979) puts it, wild tetraploid cottons are strand plants adapted to mobile shorelines, with impermeable seeds adapted to salt water diffusion. The instability of their habitat “is in itself highly stable” and very ancient, so “that the pioneers are simultaneously old residents”.
Extrapolating this TWC climatic model to South America and Polynesia points towards places where other wild representatives of tetraploid Gossypium species have been reported, which gives a strong example of niche conservatism. Thus, the distribution model has perfectly captured those conditions that are related to both biotic and abiotic constraints for the development of TWC populations.
To be continued…