- The Tatar Information Agency announces a conference to celebrate NI Vavilov’s 125th birthday, neglects to include date: 25 November 2012?
- Rice and ritual expresses the “intangible cultural heritage of agriculture and food”.
- Canadian scores four new old tomatoes from their genebank.
- The agave fight goes on.
Next-generation sequencing and genebanks: a teaser
We’re of course all holding our breath, are we not, over the imminent appearance of the American Journal of Botany Special Issue on what next-generation sequencing (NGS) technologies mean for the plant sciences. A few teasers are already out on the journal’s website, and it looks like the papers will come out in piecemeal fashion over the next weeks, and months for all I know. The paper that’s most relevant to us here is perhaps that of Susan McCouch and others on NGS and genebanks. I saw an early version of it, but am not allowed to share it, so until it comes out officially, here’s a taster from the introduction to the volume as a whole by Ashley N. Egan, Jessica Schlueter and David M. Spooner. I trust the journal will consider it fair use and not come after us with their lawyers.
A total of 1750 national and international gene banks worldwide preserve ~7 million accessions of advanced cultivars, landraces, and wild species relatives of plants that the world depends on for food, fiber, and fuel (FAO, 2010 ). McCouch et al. (2012) present a vision for the potential of large-scale genotyping to help characterize, use, and manage gene bank collections, from their perspectives as scientists working with large-scale rice collections. Genebanks have many pressing challenges due to the large size of their collections and the need to characterize them properly for a wide variety of users. They also face legal constraints (and opportunities) imposed in today’s climate of ownership of genetic resources. The challenges include the need to correctly identify accessions, track seed lots, varieties, and alleles, identify and eliminate duplicate accessions, justify adding new accessions to the collection, identify a small subset of the collection that represents a majority of the variation in the entire collection (a “core collection”), identify geographic areas holding useful sets of diverse alleles, associate genotypes with phenotypes, and motivate innovative collaborations to place useful materials into the hands of plant breeders. McCouch et al. (2012) outline these challenges and show how NGS can vastly improve genetic characterization efforts in genebanks. Initial NGS projects with the rice collections include identification of SNPs and other polymorphisms (http://www.oryzasnp. org/; http://www.ricediversity.org/; http://www.ricesnp.org/) based on large-scale resequencing and genotyping projects.
Back with a full discussion (and a comparison with the paper on the same subject in a recent Brainfood) when the publication is online.
Nibbles: PGR course, Vegetable seed kits, Maize data, Rice metabolomics, Rewilding, Sheep diversity, Llama economics, Canary flora, Cuba urban ag, Ducks, Cynara, Food sovereignty
- The annual Wageningen agrobiodiversity conservation course is in India this year. Hannes says it’s really good.
- AVRDC’s veggie seed kits are a hit in Orissa.
- CIMMYT swimming in data. Waving, not drowning.
- IRRI not doing badly either. But “quantitative train loci” is a new one on me.
- Australia thinking about introducing elephants. Yeah because that sort of thing has been such a success in the past.
- Well, actually, with sheep, I suppose it has, in a way. And the genetics says breeders have a lot of diversity to play with still.
- Your mama is a llama.
- The Island of Dogs has crop wild relatives. No, not the one in London.
- Cuba goes for new urban crops. And not for the first time, Shirley. But what about the old ones?
- Quick, duck! This piece has been rather a hit for me over on Facebook.
- VII International Symposium on Artichoke, Cardoon and Their Wild Relatives: The (Very Expensive) Book.
- Food sovereignty “started in New England”. Huh? And proponents “want to eat and sell the food they grow free from interference from state and federal regulators”. Huh?
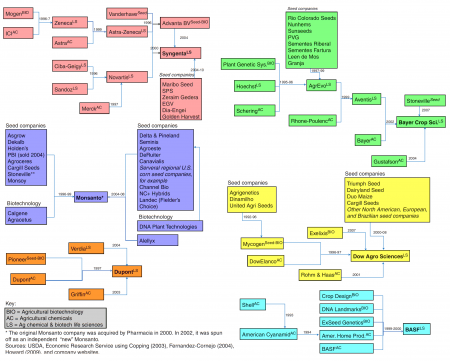
Consolidation in the seed industry
We’ve written before about consolidation in the seed industry making the same tired joke that I want to make again now, that a picture is worth 1000 words. It doesn’t hurt to stay up to date though, which is why I am grateful to Resources Research for finding, and colouring in, the diagram from the USDA report on Research Investments and Market Structure in the Food Processing, Agricultural Input, and Biofuel Industries Worldwide.
The USDA report is not, as far as I can tell, anything to do with the US Department of Justice’s investigation of antitrust practices in the seed industry, announced a couple of years ago. So what has become of that? Anyone know?
Brainfood: Falcons, Wild soybean squared, Horse domestication
- Effects of Introducing Threatened Falcons into Vineyards on Abundance of Passeriformes and Bird Damage to Grapes. Potential savings of US$234/ha for Sauvignon Blanc, more for Pinot Noir.
- Genetic characterization and gene flow in different geographical-distance neighbouring natural populations of wild soybean (Glycine soja Sieb. & Zucc.) and implications for protection from GM soybeans. There is a small amount of outcrossing, which decreases with distance. GM crops should be grown far from wild populations, certainly more than 1.5km. And we can work out better ways to collect for ex situ conservation.
- Phylogenetic relationships, interspecific hybridization and origin of some rare characters of wild soybean in the subgenus Glycine soja in China. Intermediate forms are closer to the wild than the cultivated species.
- Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. A diversity of maternal lines were domesticated about 150,000 years ago, leading to about 18 modern haplogroups. One of them is only found in the only remaining wild horse, E. przewalskii.