- “Something researchers are looking at is which cultivars, or strains, of hemp are best for the various uses — fiber, oil, nutrition, etc.” Love that etc.
- Speaking about grass: Brachiaria goes home, to wild acclaim.
- Did someone say wild? Wild grass needs help!
- Rice is a grass. Oh my yes.
- How to keep up to date with taxonomic research online.
- Pacific Regional Breadfruit Initiative gets an award.
- You can also make flour from acorns.
- And maize: what’s a grit?
- Greenpeace touts MAS.
- Next thing you know they’ll be singing the praises of Big Data. Yeah, maybe not.
The identity of “millet” in East Africa
A blog post from the Chicago Council on Global Affairs a couple of days ago reminded me that we had blogged about GYGA — the Global Yield Gap and Water Productivity Atlas — a couple of years back. Just to remind us all
…the target of the Global Yield Gap Atlas (GYGA) is to provide best available estimates of the exploitable yield gap (Yg-E) — difference between current average farm yields and 80% of Yp and Yw. Water resources to support rainfed and irrigated agriculture also are limited, which means efficiency in converting water to food, water productivity (WP), is another key food security benchmark included in the Atlas.
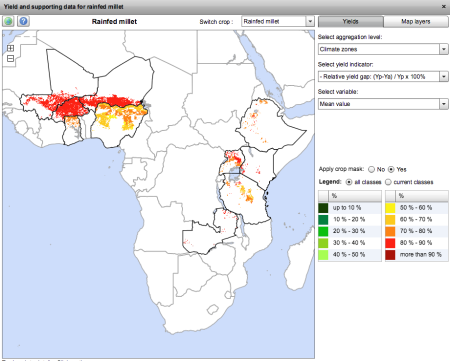
I’ve had a quick look at the GYGA website, and it seems serviceable enough, though it has the drawbacks, often remarked on in these pages, that it is difficult to share the maps you make, and import your own data into them. Here I’d like to point out another potential issue, though. This is what you get when you look at the yield gap for “rainfed millet.” Because of the aforementioned drawbacks, a clunky screenshot is the best I can manage, I’m afraid.
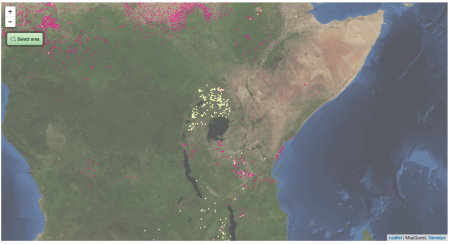
Now, the stuff in West Africa is clearly pearl millet, but what about in East Africa? Is there really that much pearl millet in Uganda, say? This is what Genesys knows about the two millets in East Africa. And yes, it’s a clunky screen grab too, but if I had wanted to, I could have downloaded separate KML files for the two crops and opened them both in Google Earth and then exported a nice JPG.
Pink is pearl, yellow is finger. There’s a little bit of pearl millet in Uganda, but not all that much. The millet there is mainly finger millet. So, which is the millet in GYGA? Is it confusing the two?
Comparing the niches of wild, feral and cultivated tetraploid cottons, Conclusion
We’re trying something new for us this week. Dr Geo Coppens co-authored an interesting paper 1 recently which brings together a number of our concerns: domestication, diversity, crop wild relatives, spatial analysis… He’s written quite a long piece about his research, which we’ll publish here in three instalments. This is the third and final instalment. Here’s the previous one.
Understanding the causes of the large difference between the distributions of TWC and that of cultivated and feral cottons is crucial for future breeding efforts. On one hand, the strong genetic divergence between wild and feral G. hirsutum that we have found in a sister SSR marker study may be partly related to differential selection and adaptation, and/or to the difficulty for domesticated forms to get established in a hostile environment. On the other hand, there is little doubt that much of the difference lies in the distinct ecological interpretation to be given to their respective models. Indeed, we may consider that the distribution of cultivated cottons approaches that of the fundamental niche of the species (i.e. where abiotic factors are favourable), which is obviously wider than the realized niche (further constrained by biotic factors, including competition).
Indeed, biotic factors are much less important under cultivation. For feral plants, we may refer to another niche concept: source-sink relations between cultivated and feral populations may explain how the latter may be found in unsuitable habitats (Pulliam, 2000). Logically, escapes from cultivation simply persist in the vicinity of cultivated cottons, in recently abandoned fields or along roadsides, benefitting from the open agricultural landscape. Why do feral populations observed further from cotton fields and dooryards, in more developed secondary vegetation (our ‘wild/feral’ category), present a very similar climatic envelope, and not an intermediate one? In fact, once interspecific competition has been avoided or reduced at the seedling stage, ancient escapes may persist for long, even in relatively high secondary vegetation. This is consistent with germplasm collectors’ reports (e.g. Ulloa et al., 2006) that feral populations are morphologically similar to local races of cultivated cotton, and that they are getting rarer as the cultivation of perennial cotton declines in Mexico.
The cotton example shows the relevance of ecological niche concepts even in the seemingly straightforward study of crop plant distribution, and particularly in the context of domestication studies, where it is crucial to distinguish between wild and feral forms. Comparing cultivation areas of domesticated forms with the original distribution of their wild relatives does not allow direct inferences on crop potential for climatic adaptation. Getting back to the tetraploid cotton example, niche conservatism after more than 1 million years of species diversification (the age of the AADD allotetraploid Gossypium genome) should make us more cautious in evaluating the result of 10,000 years of breeding on crop climatic adaptation. While there is no doubt that domestication and cultivation have resulted in widening of crops’ climatic envelopes, we need to understand much better the respective shares of genome and environment in the process. In the face of global climate change, we will need the tools of both genetic improvement and agroecology.
References
Brubaker CL, Wendel JF (1994) Reevaluating the origin of domesticated cotton (Gossypium hirsutum, Malvaceae) using nuclear restriction fragment length polymorphism (RFLP). Am J Bot 81: 1309–1326. doi: 10.2307/2445407
Coppens d’Eeckenbrugge G, Lacape J-M (2014) Distribution and Differentiation of Wild, Feral, and Cultivated Populations of Perennial Upland Cotton (Gossypium hirsutum L.) in Mesoamerica and the Caribbean. PLoS ONE 9(9): e107458. doi:10.1371/journal.pone.0107458
Fryxell PA (1979) The Natural History of the Cotton Tribe (Malvaceae, Tribe Gossypieae): Texas A&M University Press, College Station, TX. 245 p.
Hutchinson JB (1954) New evidence on the origin of the old world cottons. Heredity 8: 225–241. doi: 10.1038/hdy.1954.20
Hutchinson JB, Silow RA, Stephens SG (1947) The evolution of Gossypium and the differenciation of the cultivated cottons: Emprire Cotton Growing Corporation. 160 p.
Miller AJ, Knouft JH (2006) GIS-based characterization of the geographic distributions of wild and cultivated populations of the Mesoamerican fruit tree Spondias purpurea (Anacardiaceae). Am J Bot 93: 1757–1767. doi: 10.3732/ajb.93.12.1757
Pulliam HR (2000) On the relationship between niche and distribution. Ecology Letters 3: 349–361. doi: 10.1046/j.1461-0248.2000.00143.x
Russell J, van Zonneveld M, Dawson IK, Booth A, Waugh R, et al. (2014) Genetic Diversity and Ecological Niche Modelling of Wild Barley: Refugia, Large-Scale Post-LGM Range Expansion and Limited Mid-Future Climate Threats? PLoS ONE 9(2): e86021. doi:10.1371/journal.pone.0086021
Sauer JD (1967) Geographic reconnaissance of seashore vegetation along the Mexican Gulf coast; Mc Intire WG, editor. Baton Rouge. 59 p.
Stephens SG (1965) The effects of domestication on certain seed and fiber properties of perennial forms of cotton, Gossypium hirsutum L. The American Naturalist 94: 365–372. doi: 10.1086/282377
Thomas E, van Zonneveld M, Loo J, Hodgkin T, Galluzzi G, et al. (2012) Present Spatial Diversity Patterns of Theobroma cacao L. in the Neotropics Reflect Genetic Differentiation in Pleistocene Refugia Followed by Human-Influenced Dispersal. PLoS ONE 7(10): e47676. doi:10.1371/journal.pone.0047676
Ulloa M, Stewart JM, Garcia EA, Godoy S, Gaytan A, et al. (2006) Cotton genetic resources in the Western states of Mexico: in situ conservation status and germplasm collection for ex situ preservation. Genet Resour Crop Evol 53: 653–668. doi: 10.1007/s10722-004-2988-0
Wendel JF, Brubaker CL, Seelenan T (2010) The origin and evolution of Gossypium. In: Stewart JM, Oosterhuis DM, Heitholt JJ, Mauney JR, editors. Physiology of cotton. Dordrecht: Spinger, Netherlands. pp. 1–18.
Comparing the niches of wild, feral and cultivated tetraploid cottons, Part 2
We’re trying something new for us this week. Dr Geo Coppens co-authored an interesting paper 1 recently which brings together a number of our concerns: domestication, diversity, crop wild relatives, spatial analysis… He’s written quite a long piece about his research, which we’ll publish here in three instalments. This is the second instalment. Here’s the first, in case you missed it.
To investigate the origin of cultivated cotton in the New World, we also had to define the distribution of the wild precursors, which meant distinguishing True Wild Cotton (TWC) populations from feral (secondarily wild) populations. We first defined five categories: (1) TWC populations, which were described as such by cotton experts with consistent ecological and/or morphological criteria; (2) wild/feral cottons, reported to grow in protected areas and/or forming relatively dense populations in secondary vegetation; (3) feral plants living in anthropogenic habitats (field and road margins); (4) cultivated perennial cottons; and (5) plants of indeterminate status. We then organized our extensive dataset of locality information from herbaria and genebanks in the same way as Russian nesting dolls.
Avoiding the problematic comparisons between consecutive categories, we first modelled G. hirsutum distribution based on the whole sample, then reducing the sample to ‘feral’+‘wild/feral’ + TWC , then to ‘wild/feral’+TWC, and finally to TWC alone. Thus, we could see that the distribution model (we used the Maxent software for niche modelling) was not significantly affected by the successive removals of specimens classified as indeterminate, cultivated, or feral. Only the model based on TWC populations diverged very clearly, their distribution being severely restricted to a few coastal habitats, in northern Yucatán and in the Caribbean, from Venezuela to Florida (Figures 1 and 2).
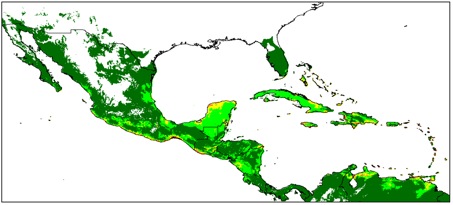
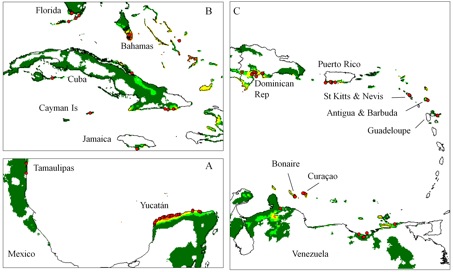
A principal component analysis on climatic parameters confirmed the absence of clear differences among the climatic niches of cultivated, feral, and ‘wild/feral’ populations, contrasting strikingly with that of TWC populations, restricted to the most arid coastal environments. The TWC habitat’s harsh conditions are quite certainly related to the fact that “Gossypium is xerophytic and that even its most mesophytic members are intolerant of competition, particularly in the seedling stage” (Hutchinson, 1954). The proximity to the sea plays a role itself, for these “extreme outpost plants” (Sauer, 1967). Indeed, as Fryxell (1979) puts it, wild tetraploid cottons are strand plants adapted to mobile shorelines, with impermeable seeds adapted to salt water diffusion. The instability of their habitat “is in itself highly stable” and very ancient, so “that the pioneers are simultaneously old residents”.
Extrapolating this TWC climatic model to South America and Polynesia points towards places where other wild representatives of tetraploid Gossypium species have been reported, which gives a strong example of niche conservatism. Thus, the distribution model has perfectly captured those conditions that are related to both biotic and abiotic constraints for the development of TWC populations.
To be continued…
Domestication and distribution: comparing the niches of wild, feral and cultivated tetraploid cottons
We’re trying something new for us this week. Dr Geo Coppens co-authored an interesting paper 1 recently which brings together a number of our concerns: domestication, diversity, crop wild relatives, spatial analysis… He’s written quite a long piece about his research, which we’ll publish here in three instalments, over the next few days days, starting today. Enjoy!
![]() Niche Modelling has been increasingly coupled with genetic studies for the study of plant domestication and/or diffusion (e.g. Thomas et al., 2012, on cacao; Russell et al., 2014, on barley). However, there has been relatively little interest in the effects of domestication and cultivation on distribution. Comparing the distributions of wild and cultivated hog plums in Meso- and Central America, Miller and Knouft (2006) found that the crop’s environmental range was significantly wider than that of its wild relatives observed in the dry forests, which was suspected to result from selection and attendant adaptation under domestication.
Niche Modelling has been increasingly coupled with genetic studies for the study of plant domestication and/or diffusion (e.g. Thomas et al., 2012, on cacao; Russell et al., 2014, on barley). However, there has been relatively little interest in the effects of domestication and cultivation on distribution. Comparing the distributions of wild and cultivated hog plums in Meso- and Central America, Miller and Knouft (2006) found that the crop’s environmental range was significantly wider than that of its wild relatives observed in the dry forests, which was suspected to result from selection and attendant adaptation under domestication.
This interpretation seems related to a combination of common ideas about species habitats, distribution modelling and the effects of human selection and diffusion on crop adaptation. Most correlative species distribution modelling has been based on the use of abiotic environmental variables, and the best documented are climatic ones. As a result, many authors think of their models in terms of climatic envelopes corresponding to the physiology and phenology of species, and remain cautious about their capacity to capture biotic effects. Therefore, it appears logical to relate the wider crop environmental range, as compared to their wild relatives, to adaptive selection under/after domestication.
Results obtained on perennial cotton from Mesoamerica and the Caribbean underline the interest of a more ecological interpretation (Coppens d’Eeckenbrugge and Lacape, 2014). Among the four independently domesticated cotton species, Gossypium hirsutum L. is by far the most widely cultivated, even in warm temperate conditions, at latitudes of up to 30°. It shares its AD allotetraploid genome with the other American domesticated cotton, G. barbadense L., native to Ecuador and Peru, and three wild species, G. darwinii Watt (Galapagos), G. mustelinum Miers ex Watt (Northeastern Brazil), and G. tomentosum Nutt. ex Seem. (Hawaii). The absence of American diploid species bearing the A genome led Hutchinson et al. (1947) to the conclusion that the A parent was an African domesticate brought by man, implying that the first allotetraploid cottons were cultivated.
On the contrary, Fryxell (1979) and Sauer (1967) recognized wild populations of G. hirsutum in Yucatán and various localities in the Caribbean. The existence of these “truly wild cottons” (TWC) was progressively substantiated by their particular seed traits (Stephens, 1965) and the isozyme study of Brubaker & Wendel (1994), indicating that the TWC populations of northern Yucatán were the likely ancestors of Mesoamerican cultivated races. More recently, molecular genetics supported Fryxell’s views, confirming that all five allotetraploid cotton species originated from a single hybridization event that took place during the Pleistocene, some 1-2 million years ago (Wendelet al. 2010). However, no systematic studies were undertaken and TWC populations were very poorly represented in subsequent studies of tetraploid cotton genetic resources. In particular, the status of TWC populations from the Caribbean was not clarified. A major practical problem was the difficulty to distinguish TWC populations from feral (secondarily wild) populations.
To be continued…