Is genebank data having a moment? Well, it’s a pretty big thing that the botanic gardens community have basically said that they need a Genesys too, and in a hugely co-authored “Perspective” article in Nature Plants to boot.
Here we have focused on the living collections data ecosystem, because many aspects to managing these collections are unique within the broader collections sector. But we can look to the more advanced and better-networked accession-level data systems of ex situ agricultural gene banks (for example Genesys), not only for inspiration but with a view to lessons learned, and ultimately as future partners in building an even broader integrated global system for ex situ conservation resources.
Hey, we’re just sitting here waiting for you to decide to join us.
And there’s other stuff going on too. The Australian Virtual Seed Bank Portal has had an update and looks just great, for example. And the Old Vine Registry‘s database has passed 4000 entries. Ok, that covers old vineyards rather than genebank collections, but same difference, don’t @ me. Anyway, would love to see it mashed up with the European Vitis Database one day.
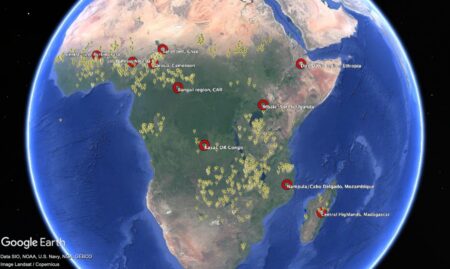
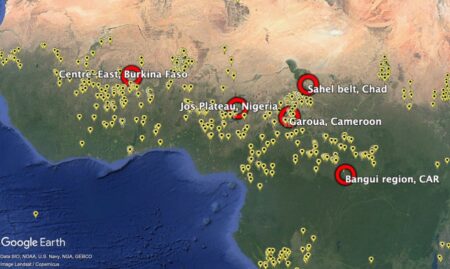
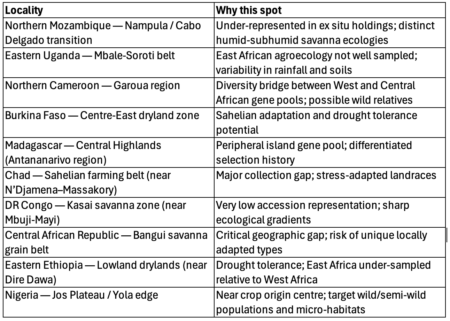
But back to genebanks. People are even building interfaces to their data, to get cool visualizations. And they’re analyzing the data to get a handle on the composition of collections , to develop monitoring and evaluation frameworks , and to identify gaps and challenges . And yes, that includes challenges in data management itself.
Documentation and data management systems required more attention in all genebanks in the study, with no genebank having full passport and inventory data in a searchable data management system, although minimum passport data on 82% of accessions was publicly available in searchable databases, including WIEWS and Genesys. Good quality, well-managed and searchable data on genebank operations are important for accurate and timely decision-making. A common issue was a backlog of data entry, with eight of the genebanks in the study relying to some extent on data retained in paper copy, field books and data sheets. Searching for accession-level data is time-consuming, and paper data sheets may be lost or damaged. Data were also stored in Excel files, making it difficult to query, or genebanks had their own customized data management system that required external support to resolve problems or make improvements. Engels and Ebert (2024) recognized the weak information management systems and online accessibility of accession-level data in national genebanks as a challenge to rationalization, as well as to cost-efficient and effective conservation and use. Despite the increasing use of digital object identifiers for accessions to link accession-level data, the Third Report on the State of the World’s Plant Genetic Resources concluded that progress in the area of documentation has been limited, and training is needed for data managers and genebank managers to adopt available improved data management systems (FAO 2025c).
So, we still have work to do. But imagine what we could achieve if we teamed up with the botanic gardens. And herbaria for that matter.