Genesys has some cool new functionality: the Subsetting Tool. With it, you have the power to create customized subsets based on the climate and soil variables of your choice. You can read more about it in a blogpost, and in more detail in the user guide. I’l probably blog about it here in due course. Any requests?
The latest on the spread of agriculture in Eurasia
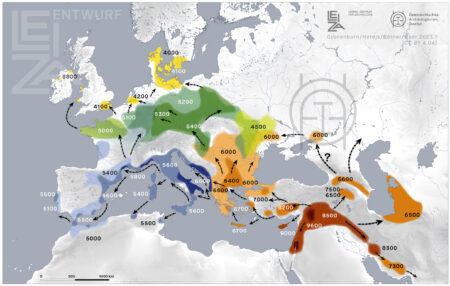
Remember the map of the spread of agriculture in Europe that I mashed up with barley genebank accessions a while back? Well, there’s a new version out, according to a tweet from Detlef Gronenborn.
It will eventually make its way here with the previous versions.
Nibbles: Calabrian citron, Cherokee seeds, Indigenous food systems & community seed bank in India, NZ apple diversity, Climate funding for food
- Calabrians are growing citrons under solar panels to protect them from the heat.
- The Church of the Good Shepherd in Decatur, Alabama is growing seeds from the Cherokee Nation Seed Bank in its gardens, and that’s a sort of homecoming.
- The Indigenous agrobiodiversity and food systems of Meghalaya in India’s NE are helping local people cope with extreme climatic conditions.
- Elsewhere in India, there are community seed banks to help with resilience.
- And in New Zealand, the Volco Park Cultivar Preservation Orchard is helping academics teach about crop diversity.
- Meanwhile, only 3% of climate funding is going into food systems.
Nibbles: Transformation, MAHARISHI, Pastoralists and climate change, Utopian okra, Landrace breeding, Ghana genebank, Indian community seedbank, Rice pan-genome, Perennial rice
- Towards resilient and sustainable agri-food systems. Summary report from the FORSEE Series of Töpfer Müller Gaßner GmbH (TMG). Take home message: We need an internationally agreed framework for agri-food systems transformation that reduces the externalities of the current systems. But how?
- Chair Summary and Meeting Outcome of the G20 Meeting of Agricultural Chief Scientists 2023. “We highlight the importance of locally adapted crops for the transition towards resilient agriculture and food systems, enhancing agricultural diversity, and improving food security and nutrition.” And that includes the wonderfully named Millets And OtHer Ancient GRains International ReSearcH Initiative (MAHARISHI). Ah, so that’s how.
- Are pastoralists and their livestock to blame for climate change? Spoiler alert: It’s complicated, but no. And here’s a digest of resources from the Land Portal explaining they can be part of sustainable and resilient agri-food systems.
- The Utopian Seed Project is developing more climate-resilient okra in the southern USA.
- Joseph Lofthouse, Julia Dakin, Shane Simonsen and Simon Gooder — interviewed here about landrace-based breeding — would approve of utopian okra.
- Plenty of landraces in the Ghana national genebank, according to this mainstream media article.
- Also plenty of landraces in India’s community seedbanks.
- Professor Zhang Jianwei at the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University has built an rice pan-genome database based on 16 (landraces presumably) accessions representing all the major sub-populations. The technical details are here. Rice sustainability and resilience no doubt beckons. Okra next?
- No, perennial rice next, apparently.
Brainfood: Pollinator evolution, Pollinator diversity, Livestock, Yak milk consumption, Poultry in situ conservation, Soil stress, Self-domestication, Natural history collections
- The expansion of agriculture has shaped the recent evolutionary history of a specialized squash pollinator. The genetic diversity of an insect crop pollinator has been affected by the fact that it pollinates a crop.
- Native pollinators improve the quality and market value of common bean. The diversity of native insect crop pollinators affects the value of the crop they pollinate.
- A global approach for natural history museum collections. Basically amounts to “ask curators what they have.” Including presumably specimens of insect pollinators. We’ve been doing this for PGRFA for quite a while now, one way or another. Back to mainly plants next week, hopefully, but let’s keep going with animals for now, and let’s see what more we can learn.
- A 12% switch from monogastric to ruminant livestock production can reduce emissions and boost crop production for 525 million people. Ruminants are not all bad after all.
- Permafrost preservation reveals proteomic evidence for yak milk consumption in the 13th century. The Mongols thought this particular ruminant was just great.
- The self-management organization as a way for the in situ conservation of native poultry genetic resources. In response to the promotion of exotic commercial poultry breeds, women’s groups in Mexico have got together and developed rules to protect native hens. Please let not these be among the 12% of monogastrics that get replaced by yaks.
- Increasing the number of stressors reduces soil ecosystem services worldwide. It’s the number of different stressors, more than their aggregate strength, that most affects how badly soils are stressed. Goes for me too, to be honest.
- Elephants as an animal model for self-domestication. I’ll believe it when elephants domesticate yaks.