- Tritodendrum hits the market.
- DS Athwal would have approved. RIP.
- Medieval bakers too, I bet.
- Want some cherry tomatoes on your bread? No? Try these then.
- Lots of crop wild relatives among newly discovered plants. See (some of) them on the new State of the World’s Plants report from Kew. And no, Kew, none of them are “miserable.”
- Early farmers unintentionally produced vegetables with larger seeds simply by cultivating them. And cereals too.
- Head of IUCN Red Data List Unit in impassioned plea for IUCN Red Data List process.
- What is taro good for? I’m glad you asked.
- I missed International Hummus Day? How could this happen?
- Textilia Linnaeana! What do you mean I’ve just had my birthday?
- Lowering the glycemic index of rice for the Chinese market.
- Fighting for avocados. Literally.
Brainfood: Canola model, Saline dates, High rice, Perennial wheat, European cowpea, Mesoamerican oil palm, Seed viability, Citrus identity, Poor cassava, Horse domestication, Wild tomatoes, Tea genome, Veggie breeding, Classical brassicas
- Development of a Statistical Crop Model to Explain the Relationship between Seed Yield and Phenotypic Diversity within the Brassica napus Genepool. Look for primary raceme area.
- Screening of Date Palm (Phoenix dactylifera L.) Cultivars for Salinity Tolerance. In Oman, Manoma and Umsila are particularly tolerant.
- Genetic structure and isolation by altitude in rice landraces of Yunnan, China revealed by nucleotide and microsatellite marker polymorphisms. Geneflow is horizontal, adaptation vertical.
- Breeding Perennial Grain Crops Based on Wheat. By adding a wild genome.
- European cowpea landraces for a more sustainable agriculture system and novel foods. 24 of them, no less. But you have to start somewhere, I suppose.
- Genetic diversity of Elaeis oleifera (HBK) Cortes populations using cross species SSRs: implication’s for germplasm utilization and conservation. From 532 palms in 19 populations to 34 individuals.
- Large-Scale Screening of Intact Tomato Seeds for Viability Using Near Infrared Reflectance Spectroscopy (NIRS). Good-bye germination tests?
- Genetic identification of ‘Limau Kacang’ (Citrus sp.), a local mandarin cultivated in West Sumatra by sequence-related amplified polymorphism (SRAP). It’s a ponkan.
- Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Cassava is decaying genetically, but breeders are helping.
- Ancient genomic changes associated with domestication of the horse. The ancient DNA of immediate post-domestication horses suggests that the stallion bottleneck happened later.
- Genetic Diversity and Population Structure of Two Tomato Species from the Galapagos Islands. They mirror island formation.
- The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. It evolved caffeine independently of coffee but not cacao. And flavour is down to a whole genome duplication.
- The contribution of international vegetable breeding to private seed companies in India. It can still make one, but for traits rather than varieties.
- Domestication, diversity and use of Brassica oleracea L., based on ancient Greek and Latin texts. Their use as a hangover cure has a long and august pedigree. No word on their raceme area.
Nibbles: Rice in Trinidad, Sweet potatoes in Ethiopia, EU crop diversity double, Sir Peter on the ginkgo, Forages, Brazilian peanuts, Seed moisture, Phenotyping double, Svalbard deposit, CATIE data, Herbarium double, Seed #resistance, Father of the apple, Agave congress
- African rice out of Africa, and out of the genebank.
- The other way around for sweet potato.
- Markers for micronutrients. And diversity for taste…
- The deep history of the ginkgo.
- Study plants to decrease the effects of burping.
- Brazil rationalizes peanut collections.
- Measuring moisture in seeds.
- Big data for better seeds. And not only in Iowa.
- NZ seeds in Svalbard.
- CATIE on Genesys.
- Oz biosecurity fail. Better stick to online.
- I want to be a seed rebel too.
- Maybe in Kazakhstan?
- Agave under the volcano under the cosh.
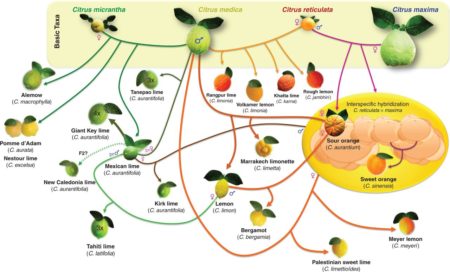
Citrus in a nutshell
Look, I suppose I could try and explain that recent Citrus phylogeny paper in Annals of Botany. But am I really likely to do better than this graphic?
I didn’t think so.
Important to know what’s what. But if all that’s just too much for you, wind down by listening to Helena Attlee on Eat this Podcast on citrus in Italy.
Brainfood: Cannabis roundup, Citrus genomes, Mapping Africa, Maize diversity, Qat diversity, Language diversity, Apple taste, Coconut diversity, Napier grass review, Rangeland management, Chinese goats, Arabica evaluation, Bangladeshi chickens, Seed endophytes
- Cannabis Domestication, Breeding History, Present-day Genetic Diversity, and Future Prospects. The weed is in trouble, man. Just one of the papers in a very special issue of Critical Reviews in Plant Sciences. And for a historical perspective…
- Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Apomixis is down to a “miniature inverted-repeat transposable element insertion in the promoter region of CitRWP.”
- Automated cropland mapping of continental Africa using Google Earth Engine cloud computing. Croplands increased by 1 Mha/yr between 2003 and 2014.
- Genetic diversity and population structure of native maize populations in Latin America and the Caribbean. Three groups: Mexico + southern S. America, lowland Mesoamerica + Caribbean, Andes.
- Phylogeography of the wild and cultivated stimulant plant qat (Catha edulis, Celastraceae) in areas of historical cultivation. Centres of origin in Kenya and Ethiopia, with limited movement between the two, but some hybrids in N. Kenya; the Yemeni stuff came from Ethiopia.
- Process-based modelling shows how climate and demography shape language diversity. If you assume human groups split after reaching a certain population size, and rainfall limits population density, you can predict language diversity in Australia.
- Volatile Compound Profiles of Malus baccata and Malus prunifolia Wild Apple Fruit. Look at the esters.
- Genetic diversity, population structure and association analysis in coconut (Cocos nucifera L.) germplasm using SSR markers. I can’t see much new here. Please, coconut experts, tell me what I’m missing.
- Opportunities for Napier Grass (Pennisetum purpureum) Improvement Using Molecular Genetics. Could do better.
- Heterogeneity as the Basis for Rangeland Management. Gotta mix it up.
- Analysis of genetic diversity of Chinese dairy goats via microsatellite markers. The locally developed breeds are all derived from a single European breed.
- Variation in bean morphology and biochemical composition measured in different genetic groups of arabica coffee (Coffea arabica L.). You can’t use morphology to predict taste.
- Breeding for the improvement of indigenous chickens of Bangladesh: evaluation of performance of first generation of indigenous chicken. I don’t understand this much, but I liked the names of the genotypes: Naked Neck, Hilly and Non-descript Desi.
- Persistence of endophytic fungi in cultivars of Lolium perenne grown from seeds stored for 22 years. It’s a record!