- Against the grain? A historical institutional analysis of access governance of plant genetic resources for food and agriculture in Ethiopia. Culture, economics and politics.
- Early Pastoral Economies and Herding Transitions in Eastern Eurasia. Everything changed around 1200 BC. Starting in Mongolia.
- Genetic diversity within and between British and Irish breeds: The maternal and paternal history of native ponies. Diversity within breeds being maintained, global haplotypes well represented, but a couple of breeds pretty unique. Long way from Mongolia.
- Diversity buffers winegrowing regions from climate change losses. Gotta change your cultivars.
- Contribution de la biodiversité à l’éco-oenotourisme des vignobles héroïques: atouts et perspectives. You can’t change your cultivars.
- Marker-assisted selection in a global barley (Hordeum vulgare subsp. vulgare) collection revealed a unique genetic determinant of the naked barley controlled by the nud locus. One genetic variant, from East Asia, makes barley naked.
- Morphological diversity within a core collection of subterranean clover (Trifolium subterraneum L.): Lessons in pasture adaptation from the wild. The Australian cultivars have similar morphological diversity to the core collection, and several morphological characters are probably adaptive.
- Genome-wide genetic diversity is maintained through decades of soybean breeding in Canada. After an initial decline, though, and there’s more out there.
- Evaluation of genetic diversity, agronomic traits, and anthracnose resistance in the NPGS Sudan Sorghum Core collection. 10% country subset of a 10% core subset of >40,000 accessions contains multiple anthracnose resistance sources, and lots of other diversity.
- Phylogeny and conservation priority assessment of Asian domestic chicken genetic resources. 7 clades, 3 centres of origin, northern Yunnan the highest priority for conservation.
- European and Asian contribution to the genetic diversity of mainland South American chickens. Alas, no evidence of a pre-Columbian Polynesian contribution. Yunnan, that’s another story.
- Arbuscular mycorrhizal fungi affect the concentration and distribution of nutrients in the grain differently in barley compared with wheat. Differently as in opposite directions.
- Molecular and Morphological Divergence of Australian Wild Rice. Including a putative new taxon.
Not that kind of flesh
Oh, Twitter, you’re such a tease.
Knowing that I am currently working on orange-fleshed sweetpotatoes, Luigi kindly sent me a link to a tweet. This one:
That red ellipse? I’m drawing your attention to Twitter’s warning that seeing the images might bring on an attack of the vapours in highly-attuned personalities.
Is it just the word “fleshed”? I had to know.
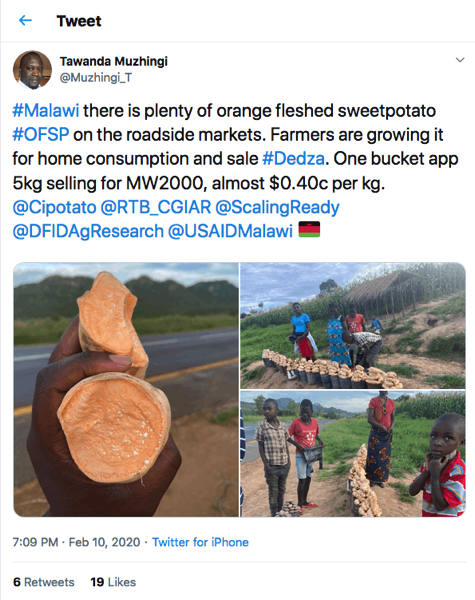
I dunno. “Filth,” they say, “is in the mind of the beholder” and I have to say, I’m not seeing it.
Orange-flesh, though. Where else have I seen that? Maybe that’s what Twitter is trying to warn me against.
No matter. Congratulations to @CIPotato and @RTB_CGIAR.
Sweet poster on bitter gourds from WorldVeg
 Had no idea Asian countries were so particular about their bitter gourds. About 500 accessions in the world’s genebanks to play with.
Had no idea Asian countries were so particular about their bitter gourds. About 500 accessions in the world’s genebanks to play with.
Help improve PGRFA data
A couple of short surveys for you, if you’re into that kind of thing.
- Help the Crop Trust improve Genesys.
- Help the Plant Treaty improve the management of data on in situ conservation of crop wild relatives.
Twenty minutes tops to do both, I promise.
Brainfood: Agrobiodiversity Index, Breeding strategy, Soybean breeding, Red Listing, Stunting, Planetary boundaries, ITPGRFA, Wheat domestication, Anthropogenic fire double, Japonica diversity, Rice landraces, Tepary breeding, Lupin genome, Hazelnut diversity, Lapita food
- Text Mining National Commitments towards Agrobiodiversity Conservation and Use. Fancy maths cannot find evidence of country commitment to seed diversity.
- Optimized breeding strategies to harness Genetic Resources with different performance levels. How a public breeding programme can help out private breeding programme.
- Introgression of novel genetic diversity to improve soybean yield. Public breeding programme helps out private breeding programme. I suppose both got something out of it.
- Rapid Least Concern: towards automating Red List assessments. Nifty web application takes all the fun out of red listing. We talked about this, people.
- Mapping child growth failure across low- and middle-income countries. Even countries and regions that are generally doing well have stubborn hotspots.
- Feeding ten billion people is possible within four terrestrial planetary boundaries. Feeding, but not necessarily nourishing.
- Genebank Operation in the Arena of Access and Benefit-Sharing Policies. Use the SMTA for everything.
- Multiregional origins of the domesticated tetraploid wheats. Semi-domesticated in the southern Levant, then moved to the northern Fertile Crescent to be finished off. Compare and contrast with barley.
- Conservation implications of limited Native American impacts in pre-contact New England. Native Americans didn’t manage woodland by controlled burning after all…
- Global change impacts on forest and fire dynamics using paleoecology and tree census data for eastern North America. …Sure they did. Interesting discussion on this on Twitter.
- Multiple streams of genetic diversity in Japonica rice. It’s basically a pan-genome.
- Genomic analyses reveal selection footprints in rice landraces grown under on‐farm conservation conditions during a short‐term period of domestication. Some interesting genetic changes after 27 years of on-farm management, but no erosion.
- Breeding tepary bean (Phaseolus acutifolius) for drought adaptation: A review. You need other species.
- High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Winter and spring varieties are genetically distinct from each other, and from landraces.
- Genetic diversity and domestication of hazelnut (Corylus avellana L.) in Turkey. Hardly domesticated at all.
- Exploitation and utilization of tropical rainforests indicated in dental calculus of ancient Oceanic Lapita culture colonists. Including bananas.
- Benchmarking genetic diversity in a third-generation breeding population of Melaleuca alternifolia. There’s still quite a bit of diversity around.