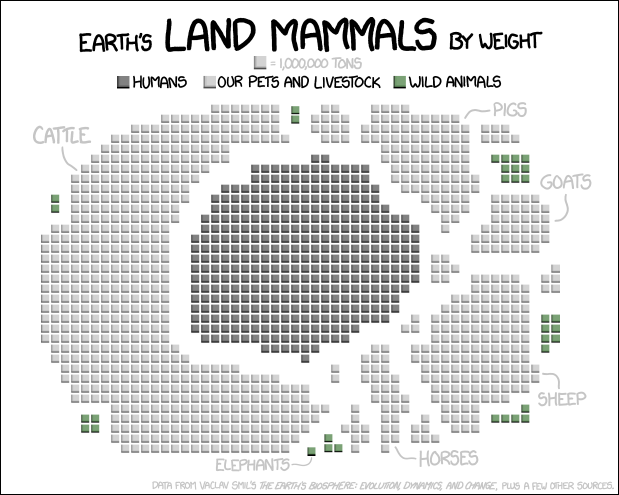
I just think you should see this, from my favourite comic on the web.
Agrobiodiversity is crops, livestock, foodways, microbes, pollinators, wild relatives …

I just think you should see this, from my favourite comic on the web.
An item in Monday’s Brainfood prompted Brian Ford-Lloyd to revisit the concept of core collections. The paper looked at “similarity groups” in genebank holdings.
One important question they addressed was ‘why identify similarity groups?’ (not to be confused with duplicates), and only time will tell whether their expectations will be met.
However, there are other issues that occurred to me. One is the relationship with ‘core collections’ (which are not mentioned in the article) of which there are now many, even for a single crop such as rice, and which are proven to be of considerable use (see: Genetic resources and conservation challenges under the threat of climate change, Ford-Lloyd, Engels and Jackson – in Plant genetic resources and climate change – Jackson, Ford-Lloyd and Parry, 2014) (sorry for the plug!). So, having identified similarity groups, is it now necessary to go back and redesign core collections? This seems unlikely, but it would perhaps be worthwhile checking core collections to see the extent of occurrence of ‘similar’ accessions. This might have particular value, not necessarily to ensure maximised diversity within core collections. It might be useful to look for similar accessions to those that have already proved to be of value within core collections, possibly revealing similarly adapted accessions of even greater value.
Some great photos of the Genetic Resources Centre Annual Field Day over on IRRI’s Flickr site. IRRI’s breeders are invited to visit the genebank’s seed multiplication plots every year, to see if anything grabs their eye. The chessboard effect is due to sowing early and late varieties in an alternating pattern. I’m assured the contrast “is becoming more pronounced each year as the supporting data improve.” See for yourself by comparing with previous years.

Speaking of Amanda, she also recently went on the Living with the Land boat ride at the Epcot theme park at Walt Disney World in Florida.
A relaxing 13-minute boat ride takes you on an informative journey through a tropical rain forest, an African desert complete with sandstorm, and the windswept plains of a small, turn-of-the-century family farm. Guests experience the struggles of the past and plans for farming in the future including Hydroponics, Aeroponics and Aquaculture. It’s not just about fruits and veggies, fish farms are on display. Since The Land is a Disney restaurant supplier, You could very well be seeing your entree. Wonder where those Mickey shaped cucumbers in your salad came from? This is where they’re grown. The educational content on this ride is geared more towards adults, but younger guests will love the boat ride and spotting the different fruits and vegetables.

Very educational, I’m sure. Anyway, this photo of hers featuring Okinawa Spinach caught my eye, even more than the Mickey shaped cucumbers, because I’d never heard of the stuff. Turns out to be Gynura bicolor, and to have really few accessions in the world’s genebanks. I wonder why Disney World picked on it in preference to any number of better known Asian vegetables. And whether they sell seeds in the gift shop. But it’s certainly one way to stimulate interest in a neglected species.