Bill Gates had a short Q&A with a BBC interviewer and a couple of African farmers on the World Service’s The World Today this morning. You can of course listen to him on iPlayer, but only for the next week or so. His bit starts at about the 13:30 mark and lasts about 10 minutes. But you can also listen to him here on our blog, where you don’t have to fiddle with the cursor and we promise not to take him down. Mr Gates does a pretty good job of sidestepping a question on slash-and-burn, and dealing head-on with one on GMOs. The project on breeding stress-tolerant rice he mentions is one the Foundation has with IRRI. But would it have killed him to mention that all breeding is underpinned by genebanks?
More pain
Don’t get frustrated, there is always hope! We have now included the possibility of directly downloading the map of the recommended locations for genetic reserves in kmz, shp and tiff formats in the main webpage. This website is focused on the dissemination of information to the general public. For those interested in more detail, the webpage of each proposed site provided links to protectedplanet.net or Natura 2000 viewer where additional information concerning the protected area can be gathered including the download of the corresponding kmz file. Anyhow, the downloadable files at the main page should facilitate the job from now on.
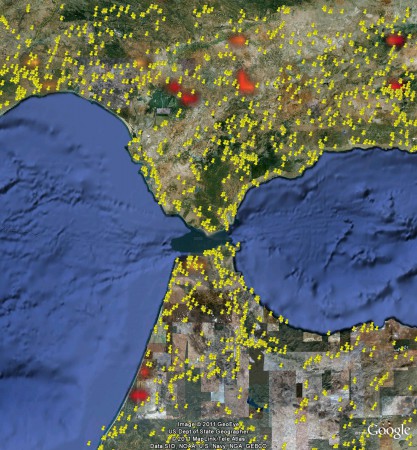
Many thanks to Jose Iriondo for this comment on my recent post on the pleasure and pain of combining biodiversity data. It’s not quite as easy as he makes out. Yes, you can download the boundaries of protected areas from the sites he mentions. This is what I got for the Estrecho site in southern Spain.
The white rectangle is all I could manage to extract from ProtectedPlanet, having gone through the pain of registering, though admittedly I’ve had better luck in the past. It could be something I’m doing wrong. The green shape I got from Natura 2000 and obviously it’s considerably better. But it still does not have the Avena, Beta, Brassica and Prunus species data in the original website AEGRO website. For that, however, it sounds like we wont have to wait for long. Jose again:
Futhermore, we will soon include the possibility of directly downloading the kmz file corresponding to each of the Google Earth plugins of each proposed site.
Hope does spring eternal.
The pleasures and frustrations of combining biodiversity data
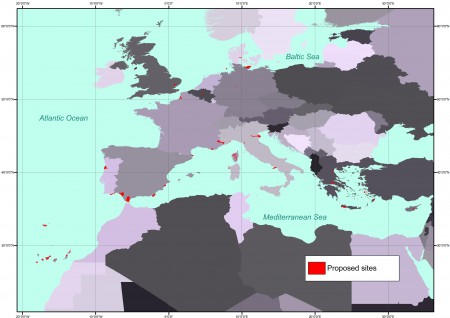
This website provides information on recommended locations, mainly in protected areas, suited for the establishment of genetic reserves for Avena, Beta, Brassica and Prunus targeted crop wild relative taxa across Europe, in the context of the AEGRO project. Available information includes ecogeographical data as well as an inventory of crop wild relatives belonging to the four target genera occurring at each location.
This is the map of the recommended sites:
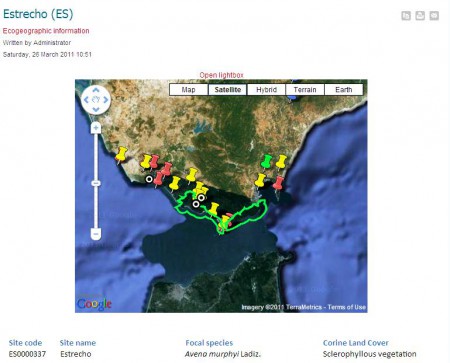
Which is great, and even greater is the fact that you can look at individual species, and the suggested protected areas, using a nifty Google Maps plugin. This, for example, is the Estrecho site in southern Spain, which is where you find an endemic wild oat (among other things).
The problem is 1 that I can find no way of mashing these data up with anything else. For example, say you want to add Genesys data to see if any other species occur in this, or any other, protected area. I don’t see how. You know where those Genesys accessions are:
But there’s no way to combine the two. Or maybe you want to see if the area was affected by fires last summer. Can’t be done. You know where the fires occurred:
But there’s no way to combine the two. Whereas of course you can easily combine that NASA fire data with the Genesys data, simply by bringing both into Google Earth. 2
So I guess my plea is: if you’re going to use Google Maps or Google Earth to display your biodiversity data, please also make it downloadable. Maybe there was a reason why this couldn’t be done in this project. I’m all ears.
Oh, and there’s another thing while I’m indulging my hobbyhorses. Can’t we use some innovative approaches to add to these kinds of datasets? I mean, if it can be done for amphibians…
Snow White and the Four Coconut Types
Over at the Coconut Google Group, Hugh Harris has had enough.
Snow White had no problem – not only was the name of each Disney dwarf carved on the end of the bed, they were identified by easily distinguished features: Happy, Sleepy, Bashful, Sneezy, Dopey, Grumpy and Doc.
Dwarf coconut names are not so simple.
- Dr Nair is looking for a source of green Malayan Dwarf
- Roland thinks there may be half a dozen green dwarf populations in Malaysia, not just one, pure green dwarf
- Dr Srinivasan calls for more insights on the distribution pattern of Malayan Green Dwarf, Brazil Green Dwarf etc.
- Elan Star wants to know about the Samoan or Tongan dwarf in Hawaii.
So perhaps this is a good time to suggest that what is needed now is a Global Coconut Genome Consortium.
For a fee, anyone should be able to send a coconut tissue sample (dwarf, tall, hybrid or unknown) to a collaborating laboratory for a DNA analysis that would identify that sample in terms of its similarity to, or difference from, all previously analysed samples.
The technology is available in many countries on all continents (except Africa?); there are already laboratories able to handle coconut samples in Australia, China, Europe, the USA and Brazil, and probably elsewhere (and the cost is coming down).
There is an International Botanical Congress in Melbourne, Australia, this July. If anyone reading this email is going to that meeting, or knows someone who will be there, please make an opportunity to talk about starting a Global Coconut Genome Consortium (GCGC or GC^2).
In the meantime, returning to the subject of dwarf coconuts, there can never be a 100% “pure” green dwarf or any other coconut (until in vitro propagation methods are a practical possibility).
Indeed, all coconut “varieties” are no more or less than local populations that look phenotypically similar to other populations that share a recent common ancestor.
At the risk of causing more, not less, confusion, I would like to suggest that the commonplace distinction of “tall or dwarf” should be forgotten and replaced. Instead we should recognise four “basic” plant habit phenotypes:
- the tall wild type (with large and small fruited sub-types)
- the tall domestic type (with different fruit-colour sub-types)
- the dwarf domestic type (with different fruit-size and -colour sub-types)
- the compact-habit dwarf (which has all the tall features except internode length)
But each of these basic phenotypes, and particularly the first two, are locally introgressed, so that there appears to be a fifth, intermediate, phenotype. For want of any accepted term, this can be thought of as the common cultivated coconut.
And, Elan, your dwarf is number 4 but if you look around you will certainly see number 3 as well.
So, a Global Coconut Genome Consortium. Any takers?
Collecting germplasm in the Iraqi desert
You may remember the story of Mrs Sanaa Abdul Wahab Al-Sheikh of the Iraqi genebank. Well, she’s still collecting away, most recently in the deserts of southern Iraq. Here are a couple of the photographs she recently sent me.