The gene for orange cauliflowers promotes storage, not manufacture, of carotenes. A new approach to enhancing nutrition?
Globalization and parasite diversity
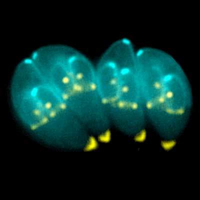 Toxoplasma gondii is a protozoan ((From H. Michael Kubisch. Photograph shows toxoplasma dividing into daughter cells. Image provided by Ke Hu and John Murray. DOI: 10.1371/journal.ppat.0020020.g001)) that can infect birds and mammals — although it can reproduce sexually only in domestic and wild cats. It has been estimated that about one in three human adults is infected, although the symptoms are usually minor. However, one particularly troublesome aspect of toxoplasma is its ability to cross the placenta from the mother and infect the growing fetus in utero. This can result in serious consequences in newborn children sometimes leading to heart and eye problems.
Toxoplasma gondii is a protozoan ((From H. Michael Kubisch. Photograph shows toxoplasma dividing into daughter cells. Image provided by Ke Hu and John Murray. DOI: 10.1371/journal.ppat.0020020.g001)) that can infect birds and mammals — although it can reproduce sexually only in domestic and wild cats. It has been estimated that about one in three human adults is infected, although the symptoms are usually minor. However, one particularly troublesome aspect of toxoplasma is its ability to cross the placenta from the mother and infect the growing fetus in utero. This can result in serious consequences in newborn children sometimes leading to heart and eye problems.
A recent study in the US has put an interesting spin on the genetic composition of toxoplasma. Toxoplasma DNA obtained from various chicken populations around the globe points to the existence of four major genetic strains, two found only in South America, one in the rest of the world — but not in South America — and a fourth population that seems to be ubiquitous.
The authors speculate that toxoplasma evolved initially in South America and then spread into Eurasia. The two populations were then separate for a long time. How this early migration might have happened is unclear; perhaps the parasite was carried by a bird. However, the spread of the Eurasian population back into North America, as well as the spread of the fourth population around the globe, could have had only one facilitator: us. At first the slave trade with its crammed and unsanitary ships possibly spread the parasite. Later, cargo ships containing agricultural goods might have given toxoplasma a lift to distant shores: in some locations the proportion of recent toxoplasma arrivals appears higher around port cities than further inland.
Yet another example of how human activity can shape evolutionary events, and contribute to diversity, in other species.
Potato foundation story
You may remember a post a few weeks back on the origins of potato late blight. Now comes news of a DNA study which looked at the origin of the European potato itself.
The spud was introduced into Europe via the Canary Islands in the mid-16th century. The authors of the study compared landraces currently grown in the Canaries, which are thought to be the descendents of those early introductions, with material from Chile and the Andes. There has long been controversy about whether European varieties trace their origins to one or the other of these places.
It turns out the answer is probably both: there were
“multiple early introductions of both Andean and Chilean germplasm to the Canary Islands and to Europe,” said Dr. David Spooner, co-author of the Crop Science study.
European aurochs DNA in domestic cattle
A study ((Edwards CJ et al., 2007. Mitochondrial DNA analysis shows a Near Eastern Neolithic origin for domestic cattle and no indication of domestication of European aurochs. Proceedings Royal Society B 274:1377-1385)) just published by the Royal Society sheds some light on the genetic relationship between the European auroch and modern European cattle breeds. Cattle were initially domesticated perhaps around 10,000 years ago in Mesopotamia, and independently in India and probably Africa. As animal agriculture spread into Europe from the Middle East, domesticated cattle must have coexisted with wild European aurochs for some time, since aurochs in Europe didn’t die out until much later (in fact, the last aurochs appear to have lived in Poland around 1627). Analysis of contemporary as well as ancient mitochondrial DNA from Middle Eastern and Central European archaeological sites now seems to suggest that European cattle originated solely from Middle Eastern aurochs, and that no introgression of European auroch genes into domesticated cattle occurred during their long coexistence.
However, an earlier study ((Götherström A et al., 2005. Cattle domestication in the Near East was followed by hybridization with aurochs bulls in Europe. Proceedings Royal Society B 272:2345-2350)) did show that there had indeed been introgression of auroch Y chromosomes into Northern and Central European domestic cattle and that these Y chromosome markers still exist in some European breeds. Of course, what might seem contradictory really isn’t: mitochondria are strictly inherited from one’s mother, and the mating of domestic cows with auroch bulls wouldn’t have left any mitochondrial evidence. It would be interesting to know whether such hybridization occurred surreptitiously or intentionally, which of course would suggest that early framers knew something about the benefits of cross-breeding.
From H. Michael Kubisch.
Undoing millennia of barley selection
Generations of beer-loving farmers have bred seed dormancy almost entirely out of barley, so that the grains will readily germinate in the malthouse. Unfortunately, that means that malting varieties are sometimes prone to jumping the gun and sprouting before harvest, while the crop is still standing in the field. That means that the grain cannot be used to make beer. Not a good thing.
Fortunately, a PhD student in Australia, a land well known for its love of the amber nectar, has compared the barley genome with that of Arabidopsis and identified some bits which may contain previously unknown dormancy genes. Should a negative effect on pre-harvest sprouting be confirmed in the field – and trials are under way – breeders could use markers for these genes to help them select genotypes which will only sprout where it would do the most good: in the maltings.