- The risks of success in quality vegetable markets: Possible genetic erosion in Marmande tomatoes (Solanum lycopersicum L.) and consumer dissatisfaction. Market takes the fun out of landraces.
- Genetic variability of root peel thickness and its influence in extractable starch from cassava (Manihot esculenta Crantz) roots. Starch content depends on dry matter content and peel thickness. About 1500 accessions from CIAT evaluated for the latter, so lots to play around with.
- A meta-analysis of crop pest and natural enemy response to landscape complexity. More landscape complexity means more natural enemies. Still no cure for cancer.
- American Chestnut past and future: Implications of restoration for resource pulses and consumer populations of eastern U.S. forests. Reintroduction of blight-resistant chestnut may have some weird effects on other species.
- Keep collecting: accurate species distribution modelling requires more collections than previously thought. Oh damn.
- Variation in seed dormancy quantitative trait loci in Arabidopsis thaliana originating from one site. Is due to two QTLs. Also flowering time. But no, I don’t understand that “In contrast…” at the end of the abstract either.
- Tracking origins of invasive herbivores through herbaria and archival DNA: the case of the horse-chestnut leaf miner. Another use for old herbarium specimens: finding evidence of pests.
- Animal breeding in India – a time for reflection, and action. The reflection is that genetic improvement has stagnated, so the action needed includes better phenotypic record keeping, more attention to local diversity and community-breeding programmes.
- Performance of micropropagation-induced off-type of East African highland banana (Musa AAA – East Africa). A promising avenue for improvement, the off-types yield more better bananas, a month later.
- Pollination services in the UK: How important are honeybees? Not as important as you might think!
- The same regulatory point mutation changed seed-dispersal structures in evolution and domestication. Cabbage-family fruit development and rice shattering share the same single point mutation.
- Participatory research and on-farm management of agricultural biodiversity in Europe. By Michel “Pimpert”. That should be Pimbert. Old news, but worth mentioning.
Brainfood: Medic systematics, Fruit wine, Alfa paper, Marula diversity, Cardamon pollination, Protein, Ants, Peanuts, Truffles, Ethiopian barley, Citrus diversity, Biofuel trees, Honeybush, Czech garlic
- Genetic similarity based on isoenzyme banding pattern among fifty species of Medicago representing eight sections (Fabaceae). People are still using isozymes? I find that oddly endearing.
- Preparation and evaluation of antioxidant capacity of Jackfruit (Artocarpus heterophyllus Lam.) wine and its protective role against radiation induced DNA damage. In other news, you can make wine from jackfruit.
- Pulping and papermaking properties of Tunisian Alfa stems (Stipa tenacissima)—Effects of refining process. Yep, a paper on paper.
- Phenotypic variations in fruits and selection potential in Sclerocarya birrea subsp. birrea. There’s a lot of it.
- Pollination studies in large cardamom (Amomum subulatum Roxb.) of Sikkim Himalayan region of India. It needs a native bumblebee.
- Effect of proteins from different sources on body composition. Hard to be sure, but probably no difference between animal and plant protein. If you’re trying to lose weight, that is.
- Ants as biological control agents in agricultural cropping systems. More common than you think, but can’t be taken for granted.
- Origin of triploid Arachis pintoi (Leguminosae) by autopolyploidy evidenced by FISH and meiotic behaviour. Maybe that was they key step on the road to the edible peanut.
- The biochemistry and biological properties of the world’s most expensive underground edible mushroom: Truffles. Not just a pretty smell.
- Ethnobotany, diverse food uses, claimed health benefits and implications on conservation of barley landraces in North Eastern Ethiopia highlands. Landraces “just” liked for culinary qualities are having a hard time.
- Comparative analysis of genetic diversity in Citrus germplasm collection using AFLP, SSAP, SAMPL and SSR markers. Boys with toys.
- Tree legumes as feedstock for sustainable biofuel production: Opportunities and challenges. Pongamia pinnata is the thing, apparently, but it’ll need research. NIMBY!
- Honeybush (Cyclopia spp.): From local cottage industry to global markets — The catalytic and supporting role of research. South African bush tea a blueprint for the development of a neglected/underutilized species? Yeah, why not.
- Diversity of S-alk(en)yl cysteine sulphoxide content within a collection of garlic (Allium sativum L.) and its association with the morphological and genetic background assessed by AFLP. Czech genebank follows up Brassica genetic diversity study with one on garlic. SACS is an important end-use trait, and varies among genetic groups.
Brainfood: Benin diversity, Catalan diversity, Serbian sorghum, Flowering in barley and sunflower, Potato nutritional quality, Cacao genebank management, Potato genebank management, Caribbean cattle, Venezuelan CWR, Ecogeographic surveys, Refugia, Vegetation change, Fisheries, Botanic gardens, Crop diversity patterns, Old trees
- Diversity, geographical, and consumption patterns of traditional vegetables in sociolinguistic communities in Benin: Implications for domestication and utilization. 245 species, from 62 families, 80% wild-harvested.
- Landraces in situ conservation: A case study in high-mountain home gardens in Vall Fosca, Catalan Pyrenees, Iberian Peninsula. 39 landraces of 31 species, disappearing fast.
- Origin, history, morphology, production, improvement, and utilization of broomcorn [Sorghum bicolor (L.) Moench] in Serbia. Summarizes 60 years of experience.
- The timing of flowering in barley and sunflower dissected. In the former, variation in photoperiod sensitivity occurred both pre- and post-domestication. In the latter, variation is clinal.
- Cultivated and wild Solanum species as potential sources for health-promoting quality traits. Some of the latter are pretty good.
- Microsatellite fingerprinting in the International Cocoa Genebank, Trinidad: accession and plot homogeneity information for germplasm management. A quarter of plots were mixtures. Well that’s no good. Huge amount of stuff in this issue of PGR-CU.
- Construction of an integrated microsatellite and key morphological characteristic database of potato varieties on the EU common catalogue. So that the above doesn’t happen.
- Footprints of selection in the ancestral admixture of a New World Creole cattle breed. Lots of African and zebu blood in Guadeloupe cattle.
- Inventory of related wild species of priority crops in Venezuela. Basically a big list.
- Potential of herbarium records to sequence phenological pattern: a case study of Aconitum heterophyllum in the Himalaya. Could be used to flesh out the above kind of thing.
- Refugia: identifying and understanding safe havens for biodiversity under climate change. How to spot refugia past and future, which would be useful for the above-but-one kind of thing.
- Modelling biome shifts and tree cover change for 2050 in West Africa. Climate change leads to greening, human impact to browning.
- Comparison of modern and historical fish catches (AD 750–1400) to inform goals for marine protected areas and sustainable fisheries. Along the Kenyan coast. Amazingly, comparisons are possible, and they show a deterioration in quality and quantity.
- The biodiversity benefits of botanic gardens. They are there, despite their history with invasives, but gardens need to get their act together. Which they are doing.
- Domesticated crop richness in human subsistence cultivation systems: a test of macroecological and economic determinants. Number of crop species grown depends on latitude, habitat heterogeneity and commitment to agriculture (as opposed to foraging, herding and exchange). Can’t make up my mind if this is interesting or predictable. Maybe it is both. Would be great to apply same method to infraspecific diversity too.
- The age of monumental olive trees (Olea europaea) in northeastern Spain. Maybe over 600 years.
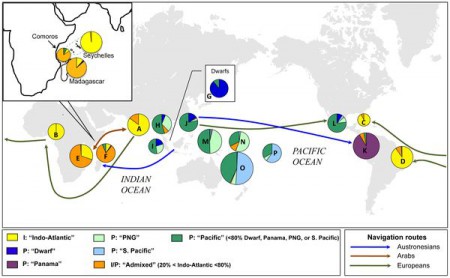
Coconut origins explained
The picture really is worth a thousand words, but if you still want the words…
Brainfood: Bean diversity, Rice domestication, Microbial interactions squared, Threat of extinction, Agroforestry, Species diversity
- Population structure and genetic differentiation among the USDA common bean (Phaseolus vulgaris L.) core collection. Subpopulations detected within usual Middle American and Andean genepools. The former was more diverse. Diversity was lower for domestication loci. One wonders whether game worth candle.
- Artificial selection for a green revolution gene during japonica rice domestication. There’s nothing new under the sun. Fuller fills us in.
- Positive plant microbial interactions in perennial ryegrass dairy pasture systems. Plant-microbe interactions can have significant positive impact on production of, and chemical inputs into and losses from, perennial ryegrass dairy pasture systems. Gotta love that agrobiodiversity.
- Plant growth promoting potential of Pontibacter niistensis in cowpea (Vigna unguiculata (L.) Walp.). And another one. New bacterium from Western Ghats fertilizes soil and helps cowpea to grow.
- Extinction risk and diversification are linked in a plant biodiversity hotspot. That would the Cape. Extinction threat (IUCN categories) is better explained by phylogeny than by human activity or plant traits. Go figure.
- The framework tree species approach to conserve medicinal trees in Uganda. Sort of like artificial keystone species. Lots of other cool stuff in the same issue of Agroforestry Systems.
- Use of topographic variability for assessing plant diversity in agricultural landscapes. By and large, more environmental variability means more plant diversity, in Switzerland. Maybe some crop wild relatives in there?