- BBC says we can feed the world through technology. And why not. The Times of India, meanwhile, didn’t get the memo.
- Bill Gates at the IFAD Governing Council will probably say the same thing today. And put his money where his mouth is.
- CIMMYT Director General said the same recently at an Economist conference. Funny how genebanks are rarely among the saviour technologies being touted.
- It’s all about the scaling up, isn’t it?
- Tell that to the Lake Chad fisherfolk who are now turning to farming.
- And, in another universe, sushi seeks protection.
- Buffel vs Rhodes in the Arabian peninsula.
Getting to the root of the Ceora family tree
Those of you who remember our little discussion here recently of grasspea breeding, and in particular the importance (or otherwise) of breeding for low values of the neurotoxin β-N-oxalyl-L-α,β-diaminopropionic acid (β-ODAP) in its seeds, will not be surprised to learn that a press release a few days ago announcing a new(ish) promising low-ODAP variety called Ceora sent me scurrying around in Genebank Database Hell.
Grass pea is a hardy annual legume with a growth habit similar to field pea, characterised by resistance to both drought and waterlogging. Its low cost, low input nature makes it a prime feed crop.
It has been restricted in use, however, due to the presence of neurotoxin ODAP, which causes paralysis of the lower limbs (lathyrism).
This was overcome by the breeding of cultivar Ceora (Lathyrus sativus) by Dr Colin Hanbury, Prof Kadambot Siddique and Dr Ashutosh Sarker of the Centre for Legumes in Mediterranean Agriculture (CLIMA).
With an ODAP level of 0.04 to 0.09 percent, Ceora virtually eliminates the toxin’s effects, making it safe for animal consumption.
Dr Gusmao has delved deeper into the plant’s benefits.
Those benefits are a suite of adaptations to cope with, and indeed also avoid, water deficits. Promising indeed. But how was this miracle plant concocted?
Ok, so googling pretty quickly revealed that Ceora, which incidentally “is the first grass pea (Lathyrus sativus) cultivar to be bred and released in Australia” …
…was derived from a cross made in 1994 at Northam, Western Australia (WA), using female parent K33 (originating from Pakistan) x male parent 8604 (originating from Bangladesh).
That’s from its official registration note. So all we now have to do is trace K33 and 8604. And that’s where I hit a wall. Because apart from finding that 8604 was also subsequently called ATC 80723 (see page 425 here), I could make no further headway. 1 Sure, 8604 might be this accession in GRIN. But then again it might not be. And of a K33 (in its various permutations) from Pakistan I could find no mention in any database. There’s a grasspea labelled K33 cv Zubryak from Russia in VIR’s database, but that’s coincidence, surely.
So I was reluctantly forced, and not for the first time, to enlist the help of a Virgil, in this case Dirk Enneking, who has commented here frequently on grasspea and other pulse breeding issues. To cut a long story short, he used his global network of grasspea contacts to discover (or re-discover, as he also found some of this stuff buried in his old notes), that 8604 is a breeding line developed by crossing the Indian line Pusa-24 (as male) with the Bangladeshi landrace “Jamalpur local” (female). Both P-24 (as it is also known) and Jamalpur local have a history of use in grasspea improvement. The former in particular is known for its low ODAP content. As for, K33 its origin seems to lie with Pakistani researcher Hafeez I.T. Khawajas, but little is know beyond that.
Anyway, all this is of any conceivable interest to anyone only if they can get hold of the germplasm. 2 Ceora is of course commercially available, although a reference sample is also stored in the USDA system. So too is P-24, although it is presumably also in the Indian genebank at NBPGR (and perhaps ICARDA, though I couldn’t find it on Genesys). Hopefully all this stuff is at Horsham, but I cannot be sure because they, like NBPGR, are not online. For now.
P.S. And here’s a final thought. This took a couple of weeks and endless emails to unravel. The much more complicated Wita 9 pedigree story just a few clicks. Once we get genebank databases sorted out, we’ll have to link them to breeders’ databases. Once.
P.P.S. Now, speaking of grain legume breeding in Australia, did someone mention the Afghan pea accession PS998?
Nibbles: Chinese agriculture, Domesticating trees, Greening economies, Genebanks, Millets
- Modern Chinese agronomist praises ancient Chinese agriculture, possibly gets in trouble.
- Domesticating trees is still the next big thing.
- Transform agriculture for a greener economy, says SciDev.net.
- VoA on genebanks, including Svalbard.
- Gerbil enthusiasts tackle millets. Yes, gerbils.
Luigi speaks! A way out of genebank database hell?
There’s nothing like being associated with manifest power to pique the interest of mainstream media. And so it was when The White House, no less, hosted a breakfast on Innovation for Global Development. Announced at the breakfast was a new version of “the Germplasm Resources Information Network-Global (GRIN-Global), a powerful but easy-to-use, Internet-based information management system for the world’s plant genebanks”. Journalists who normally wouldn’t give GBDBH the time of day were suddenly queuing to explain why this was such a good thing. And among the people they queued for was the Global Crop Diversity Trust’s Luigi Guarino. Here’s the BBC World Service’s story. Luigi speaks at 1’21”
GRIN Global on the BBC World Service, Feb 18 2012
Suitably enlightened, forgive my cynicism when I wonder what prompted a web page about the International Crop Information System to suddenly bleep on my horizon-scanning radar. At first blush it looks like it does much the same job as GRIN-Global, but that can’t possibly be correct. It must do something slightly different — powerful, undoubtedly, but by the look of things not easy to use and not internet based — which I welcome in the name of diversity and the resilience that promotes.
Who should we thank for Wita 9?
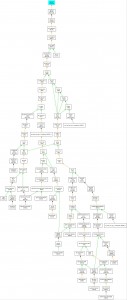 A tweet by the FAO Media Center alerted us to a feel-good story about women and rice in Côte d’Ivoire. unfortunately, I couldn’t catch the name of the variety, but the folks at FAO explained it was something called WITA 9. That turns out to be an oldish (1998) Oryza sativa variety that’s been tested quite a bit in West Africa, for example in comparison to NERICAs. With a little help, I looked it up in IRRI’s germplasm management system (see its pedigree at the side) and it turns out that those Ivorian ladies have researchers in three CGIAR Centres (IRRI, CIAT and AfricaRice), germplasm from half a dozen countries and of course the IRRI genebank to thank for their new livelihood option.
A tweet by the FAO Media Center alerted us to a feel-good story about women and rice in Côte d’Ivoire. unfortunately, I couldn’t catch the name of the variety, but the folks at FAO explained it was something called WITA 9. That turns out to be an oldish (1998) Oryza sativa variety that’s been tested quite a bit in West Africa, for example in comparison to NERICAs. With a little help, I looked it up in IRRI’s germplasm management system (see its pedigree at the side) and it turns out that those Ivorian ladies have researchers in three CGIAR Centres (IRRI, CIAT and AfricaRice), germplasm from half a dozen countries and of course the IRRI genebank to thank for their new livelihood option.