- Climate change adaptation of coffee production in space and time. There’s a plan, at least for Nicaragua.
- Journey to the east: Diverse routes and variable flowering times for wheat and barley en route to prehistoric China. Growing in diverse environments pre-adapted barley for its shift to spring sowing and move eastwards to China.
- Capturing haplotypes in germplasm core collections using bioinformatics. Fortunately, “the number of accessions necessary to capture a given percentage of the haplotypic diversity present in the entire collection can be estimated.”
- Pollinator Diversity: Distribution, Ecological Function, and Conservation. 350,000 species!
- Roots, Tubers and Bananas: Planning and research for climate resilience. Much the same, but faster.
- Priorities for enhancing the ex situ conservation and use of Australian crop wild relatives. Go north, young woman.
- Consequences of climate change for conserving leafy vegetable CWR in Europe. Go, err, northwest.
- Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Monophyletic clade which originated along the Andes of W to NW South America and spread clockwise around the Amazon.
- The Aegilops tauschii genome reveals multiple impacts of transposons. The D genome bites the dust.
Nibbles: Joanne Labate, Gebisa Ejeta, David Spooner, Strawberry 101, Mad honey, First figs, Agrobiodiversity maps, School project, Takesgiving, Private investment
- USDA vegetable crop curator tells it like it is.
- $5 million to find more Striga resistance genes in sorghum.
- Wild potato herbarium specimens find good home.
- How two New World strawberries got together in the Old World and then spread all over the world.
- Hallucinogenic honey: what could possibly go wrong?
- First farmers gave a fig.
- The other of all agrobiodiversity map mashups.
- Cool school project on crop diversity in Europe.
- In other news, “Columbusing” is a thing.
- Private sector investment in conservation: Turning “small and new” into “big and familiar.”
Brainfood: Banana diversity, Cacao and CC, Coffee and CC, Zosya diversity, Certification, Genetic surrogates, Potato diversity, Food sovereignty, Swiss wheat, Seed storage, Golden potato
- Diversity and morphological characterization of Musa spp. in North Kivu and Ituri provinces, Eastern Democratic Republic of Congo. New cultivars still being discovered.
- A review of research on the effects of drought and temperature stress and increased CO2 on Theobroma cacao L., and the role of genetic diversity to address climate change. We have the diversity. But for how long?
- Climate change adaptation of coffee production in space and time. Gonna need Plans B and C. But do we have the diversity?
- Evaluation and Breeding of Zoysiagrass Using Japan’s Natural Genetic Resources. Stick to morphology.
- Where are commodity crops certified, and what does it mean for conservation and poverty alleviation? Less for poverty alleviation than for conservation. But more and better spatial data needed, especially on organic certification.
- Environmental and geographic variables are effective surrogates for genetic variation in conservation planning. Phew!
- Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. More diversity in the landraces compared to wild species than in any other crop, few genes involved in early improvement, and different loci for adaptation to uplands and lowlands; also, wild relatives involved in diversification of long-day types.
- Agricultural biodiversity is sustained in the framework of food sovereignty. Peasants feed the world.
- Crop domestication facilitated rapid geographical expansion of a specialist pollinator, the squash bee Peponapis pruinosa. Bee follows crop follows people.
- Unlocking the diversity of genebanks: whole-genome marker analysis of Swiss bread wheat and spelt. Early breeders missed some stuff.
- A probabilistic model for tropical tree seed desiccation tolerance and storage classification. Predict storage behaviour from morphology.
- Potential of golden potatoes to improve vitamin A and vitamin E status in developing countries. Here we go again.
Seeds in motion
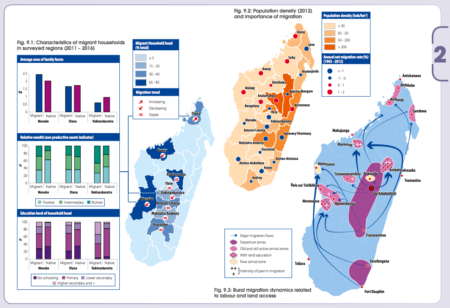
I guess it’s an occupational hazard, but whenever I see such maps, as in the new atlas Rural Africa in motion. Dynamics and drivers of migration south of the Sahara from FAO, my first question is: how many of those people are carrying seeds?
Brainfood: Rice introgression, African rice cores, Cereal domestication rates, Power vegetables, Biodiversity services, Afrikaner cattle diversity, Conservation funding
- Introgression from cultivated rice alters genetic structures of wild relative populations: implications for in situ conservation. Not totally wild any more.
- Genetic Variation and Population Structure of Oryza glaberrima and Development of a Mini-Core Collection Using DArTseq. 2,179 accessions, 5 geographic groups, 16% recover >95% of polymorphisms.
- Geographic mosaics and changing rates of cereal domestication. Applying fancy maths to archaeobotanical remains shows that selection pressures varied in time, and started slow.
- Tapping the economic and nutritional power of vegetables. Eat your veggies, damn it!
- To what extent can ecosystem services motivate protecting biodiversity? Not enough.
- Genetic diversity of Afrikaner cattle in southern Africa. 3 groups, but not geographically determined, and lots of diversity despite recent declines in numbers.
- Nominal 30-m Cropland Extent Map of Continental Africa by Integrating Pixel-Based and Object-Based Algorithms Using Sentinel-2 and Landsat-8 Data on Google Earth Engine. Next level. But when will be be able to distinguish crops?
- Reductions in global biodiversity loss predicted from conservation spending. But the impact of spending goes down with with increasing development pressure.