- Murex from Meninx.
- Breeding a better bee.
- Menswear retailer to dabble in sustainable livestock.
- There was a fourth sister.
- A world olive collection in Spain.
- “If we can assume the fire pattern is as bad as the stuff we see on TV, I reckon at this very moment there is a goodly sized handful of species that are technically extinct right now.” At least in the wild.
- Cambodia’s world-beating peppercorn makes a post-post-colonial comeback.
Brainfood: Bean diversity treble, Edited tomatoes, Breeding strategies, Pseudocereals, American lost crops, Genotyping collections, Apple diversity, Rice domestication, Cotton domestication, Wheat & gluten, Rubus phylogeny, Cassava brown streak, Prices & migration, Edible caterpillars
- Resequencing of 683 common bean genotypes identifies yield component trait associations across a north–south cline. There’s a fairly straightforward way to select for larger beans as a key component of yield.
- Is the USDA core collection of common bean representative of genetic diversity of the species, as assessed by SNP diversity? Not as much as it could be.
- Diversity, use and production of farmers’ varieties of common bean (Phaseolus vulgaris L., Fabaceae) in southwestern and northeastern Ethiopia. There are more varieties per household in places where the overall number of varieties per community was lower.
- Rapid customization of Solanaceae fruit crops for urban agriculture. Gene editing for shorter tomatoes.
- The home field advantage of modern plant breeding. Public breeding programs should go for specialist varieties that perform reliably in narrow environments.
- Thinking Outside of the Cereal Box: Breeding Underutilized (Pseudo)Cereals for Improved Human Nutrition. The next quinoa awaits its 15 minutes.
- Experimental Cultivation of Eastern North America’s Lost Crops: Insights into Agricultural Practice and Yield Potential. There’s life in the old crops yet. And that’s before gene editing.
- Time for a paradigm shift in the use of plant genetic resources. Genotype everything.
- Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. 3 early modern cultivars had a disproportionate impact on modern apples.
- Machine Learning Reveals Spatiotemporal Genome Evolution in Asian Rice Domestication. The indica and japonica sub-species have exchanged a lot of genetic material at different times, and you get different answers to the question of domestication depending on which bits you look at.
- Genetic Analysis of the Transition from Wild to Domesticated Cotton (G. hirsutum L.). There are fibre quality genes in the subgenome from the parent with unspinnable fibre. Go figure.
- A Comparative Study of Modern and Heirloom Wheat on Indicators of Gastrointestinal Health. Not much difference.
- Target Capture Sequencing Unravels Rubus Evolution. The taxonomy needs work. You don’t say.
- Expansion of the cassava brown streak pandemic in Uganda revealed by annual field survey data for 2004 to 2017. The history of a disease outbreak in excruciating detail.
- Crop prices and the individual decision to migrate. Decrease in the price of coffee in Vietnam (but not rice, which is mainly used for household consumption rather than export) resulted in increased chance of migration, but only for individuals of lower education.
- The contribution of ‘chitoumou’, the edible caterpillar Cirina butyrospermi, to the food security of smallholder farmers in southwestern Burkina Faso. It’s significant, but only during the caterpillar season. I guess they don’t keep. I spot an opportunity. Yeah, you guessed it, gene editing.
Nibbles: Legume year, Kenyan taro, Banana history, Xylella animation
- In praise of pulses.
- In praise of taro.
- In praise of a podcast on bananas.
- In praise of Hellen Mirren’s work on behalf of Xylella awareness.
Brainfood: Potato genebanks, Aichi 11, Taming foxes, Fruit diversity, Polyploidy review, Evaluating quinoa, Into Africa, IPCC review, Desiccation tolerance, Pig diversity, Oolong diversity, Wild millet, Sustainable diets
- Ex Situ Conservation of Potato [Solanum Section Petota (Solanaceae)] Genetic Resources in Genebanks. The only review of the subject you’ll need. Until the next one.
- Editorial Essay: An update on progress towards Aichi Biodiversity Target 11. Not bad, but the difficult stuff remains difficult. One of several interesting papers.
- The History of Farm Foxes Undermines the Animal Domestication Syndrome. Those Russian foxes were already pretty tame. Here’s a Twitter tread from one of the authors that lays it all out.
- Developing fruit tree portfolios that link agriculture more effectively with nutrition and health: a new approach for providing year-round micronutrients to smallholder farmers. 11 species can address micronutrient gaps.
- Plant Polyploidy: Origin, Evolution, and Its Influence on Crop Domestication. Extreme events and disasters drive polyploidy, which drives diversification at various levels, which facilitates domestication.
- Spectral Reflectance Indices and Physiological Parameters in Quinoa under Contrasting Irrigation Regimes. Phenotyping for drought tolerance from space.
- Asian Crop Dispersal in Africa and Late Holocene Human Adaptation to Tropical Environments. Via NE Africa always something new.
- Invited review: Intergovernmental Panel on Climate Change, agriculture, and food — A case of shifting cultivation and history. The IPCC could have done a better job of synthesizing the data on the impact of climate change on crops and livestock.
- Seed comparative genomics in three coffee species identify desiccation tolerance mechanisms in intermediate seeds. Whole bunch of genes involved.
- Capturing genetic diversity – an assessment of the nation’s gene bank in securing Duroc pigs. Genebank doing a pretty good job in this case.
- Genetic diversity of oolong tea (Camellia sinensis) germplasms based on the nanofluidic array of single-nucleotide polymorphism (SNP) markers. It’s not all the same.
- Tapping Pennisetum violaceum, a wild relative of pearl millet (Pennisetum glaucum), for resistance to blast (caused by Magnaporthe grisea) and rust (caused by Puccinia substriata var. indica). Out of 305 accessions, one was resistant to both diseases. IP21711 if you must know. A few more were resistant to one or the other disease.
- Can Diets Be Healthy, Sustainable, and Equitable? No, and they’ll be difficult to change, but the “burden of change should not be solely placed on the consumer’s ability to make healthy choices.”
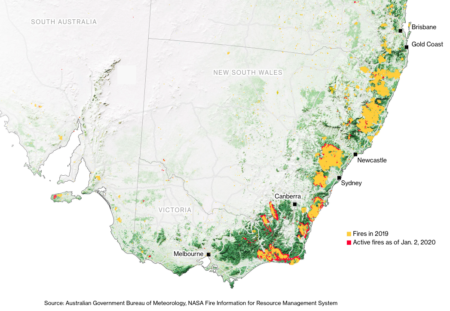
Wild sorghum burned out
Was organizing to collect an isolated population of native sorghum laeocladum at Buchanan in East Gippsland, Vic. Whole site burnt. Emphasises the importance of seedbanks for ex situ conservation of crop wild relatives #cwr @GlobalPlantGPC #sorghum #AustraliaOnFire @harrymyrans
— Prof Ros Gleadow (@RosGleadow) January 6, 2020
Indeed. Here’s the map of the distribution of Sorghum leiocladum according to GBIF.
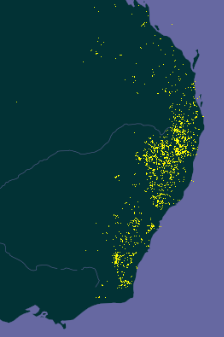
Compare with where those terrible fires are happening.
The species is in the tertiary genepool, so not particularly closely related to the crop, but there’s an indication of some potential for use in breeding for protein content.
There are 35 accessions in the genebanks that Genesys knows about, mainly in the Australian Grains Genebank. Unfortunately, I can’t find locality data. But they do seem to be in the Svalbard Global Seed Vault, although Genesys seems to think otherwise. Must look into that…