- Milk of ruminants in ceramic baby bottles from prehistoric child graves. Neolithic sippy cups. Cute.
- Heat and Drought Stress Advanced Global Wheat Harvest Timing from 1981–2014. 2.5 days per decade.
- A strong east–west Mediterranean divergence supports a new phylogeographic history of the carob tree (Ceratonia siliqua, Leguminosae) and multiple domestications from native populations. No evidence of an eastern refugium.
- Value chains to improve diets: Diagnostics to support intervention design in Malawi. You can modify existing social protection interventions to optimize diets (including increasing diet diversity) by enhancing public- and private-sector linkages.
- Contemporary evolution of maize landraces and their wild relatives influenced by gene flow with modern maize varieties. Landrace genetic diversity actually increased due to introgression from modern varieties.
- Ancient genomes reveal early Andean farmers selected common beans while preserving diversity. Because they applied weak selection. Can breeders learn from this? Also, is it similar for maize?
- Reconstruction of nine thousand years of agriculture-based diet and impact on human genetic diversity in Asia. Changes in diet through domestication and processing have left signatures on the human genome.
- Transgressive segregations for agronomic improvement using interspecific crosses between C. arietinum L. x C. reticulatum Ladiz. and C. arietinum L. x C. echinospermum Davis species. For things like pod number, earliness and tolerance to cold.
- Linking global crop and livestock consumption to local production hotspots. China is the largest consumer of primary crops, and the third largest consumer of livestock. The Corn Belt, cerrado, Europe and E. China feeds it, and the world.
- How conservation initiatives go to scale. With great difficulty.
- Morphometrics Reveals Complex and Heritable Apple Leaf Shapes. It’s mainly about aspect ratio.
- Nutrient and Antinutrient Composition of Winged Bean (Psophocarpus tetragonolobus (L.) DC.) Seeds and Tubers. The best, and worst, among 50 accessions. Spoiler alert: it depends on the nutrient, and on whether you prefer the seeds or tubers.
- Identification of Founding Accessions and Patterns of Relatedness and Inbreeding Derived from Historical Pedigree Data in a White Clover Germplasm Collection in New Zealand. 15,000 accessions trace to about 175 founders.
- Poverty not taste drives the consumption of protected species in Madagascar. Let them eat domestic livestock meat.
Rethinking reforestation
One of the reforestation papers we blogged about a few months back is coming in for some criticism.
In the original study, ecologist Thomas Crowther of the Swiss Institute of Technology in Zurich and his colleagues first used a machine learning algorithm to predict where additional trees could naturally grow, based on climatic conditions under which existing forests are known to exist. Then, his team used a handful of published estimates on the carbon stored in existing forests to estimate how much carbon those additional trees could lock in once they reach maturity. After taking into account the carbon that would be trapped in the soil, leaf litter, and dead wood associated with the trees, they arrived at their 205 gigaton estimate.
That’s a trillion trees on almost a billion hectares. Just google those figures to get an idea of the impact the paper had.
Anyway, now researchers are finding holes in the methodology. My reliably pernickety friend Eike Luedeling is objecting to the figure used to convert canopy cover to amount of carbon sequestered, and to how the availability of land for reforestation was estimated. Others are suggesting that the effect of new forests on the surface albedo should have been factored in. But perhaps the main objection is higher-level.
Several groups of scientists took particular issue with the paper’s original statement that global tree restoration is “our most effective climate change solution to date,” an assertion one of the critics called “dangerously misleading” as it implies trees are the unique solution to climate change. Land, and how we use it, can be a big part of the solution to climate change, as outlined highlighted in a recent report by the Intergovernmental Panel on Climate Change. But those strategies only “buy us time” while people cut greenhouse gas emissions, which is arguably the most powerful climate change mitigation strategy, says Luedeling.
Needless to say, the authors are countering vigorously. You can read all the toing and froing in The Scientist.
Wheat diversity for health
The objective of this conference is bringing the Wheat researchers and the Human Health community together to exchange information and discuss strategies for improving the health benefits of wheat by exploiting diverse genetic resources, modern breeding, genomics, and sustainable production and processing technologies.
You had me at Weißbier.

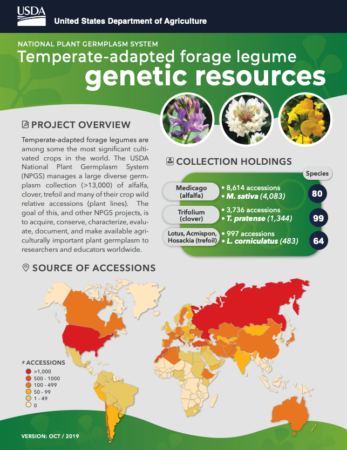
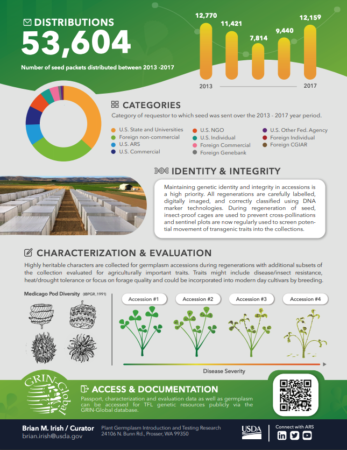
Temperate forages at the NPGS
We just got hold of a very neat infographic summarizing the work of the USDA’s National Plant Germplasm System on the Management of Temperate-adapted Forage Legume Genetic Resources and Associated Information.
If you were to scan that QR code, you’d get here, and be able to select and order material from the USDA’s GRIN-Global site.
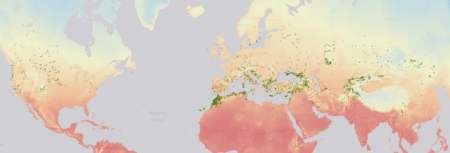
Here’s the distribution of over 6000 USDA-held accessions of Medicago, Trifolium and Lotus with geographical coordinates according to Genesys.
The colours show annual mean temperature.
Brainfood: Diversification, Wheat genomics, Historical tom, Crop mapping, African crops & CC, Trans CWR, Fish nutrition, Seed storage, Indian rice, Food Neighbourhoods, Diet sustainability, Onion evaluation, Aussie wild rice, Rice evaluation
- To diversify or not to diversify, that is the question. Pursuing agricultural development for smallholder farmers in marginal areas of Ghana. Diversify.
- Improving grain yield, stress resilience and quality of bread wheat using large-scale genomics. A genotype –> phenotype map at last. I guess that means breeders are superfluous.
- The earliest recorded tomato in Britain, in Wales. In 1590, no less.
- Biotechnology of the sweetpotato: ensuring global food and nutrition security in the face of climate change. A whole special issue. Our troubles are over.
- Probabilistic global maps of crop-specific areas from 1961 to 2014. A new, different, cooler algorithm provides somewhat different results to older, less cool algorithms.
- Potential adaptive strategies for 29 sub-Saharan crops under future climate change. Climatic conditions not currently experienced by these crops will spread, but CWRs and diversity from outside Africa might help.
- Trans Situ Conservation of Crop Wild Relatives. Just means properly integrated in and ex situ.
- Criar y Dejarse Criar: Trans-Situ Crop Conservation and Indigenous Landscape Management through a Network of Global Food Neighborhoods. See what it means? Scaling up the Parque de la Papa.
- Harnessing global fisheries to tackle micronutrient deficiencies. Small fish from the tropics could be really good for nutrition in some countries. Namibia, I’m looking at you.
- Artificial seed aging reveals the invisible fraction: Implications for evolution experiments using the resurrection approach. Store your seeds properly.
- Status of Rice (Oryza sativa L.) Genepool Collected from Western Ghats Region of India: Gap Analysis and Diversity Distribution Mapping using GIS Tools. Out of 678 rice landraces from this region, 43 have been used in crop improvement.
- Advancing an Integrative Framework to Evaluate Sustainability in National Dietary Guidelines. In 32 sub-dimensions, no less. Important.
- Assembly and characterisation of a unique onion diversity set identifies resistance to Fusarium basal rot and improved seedling vigour. Group according to local daylength.
- Australian wild rice populations: a key resource for global food security. Because they’ve been isolated from the crop.
- Novel method for evaluation of anaerobic germination in rice and its application to diverse genetic collections. No word on whether it’s applicable to Aussie wild species, but I bet it is.