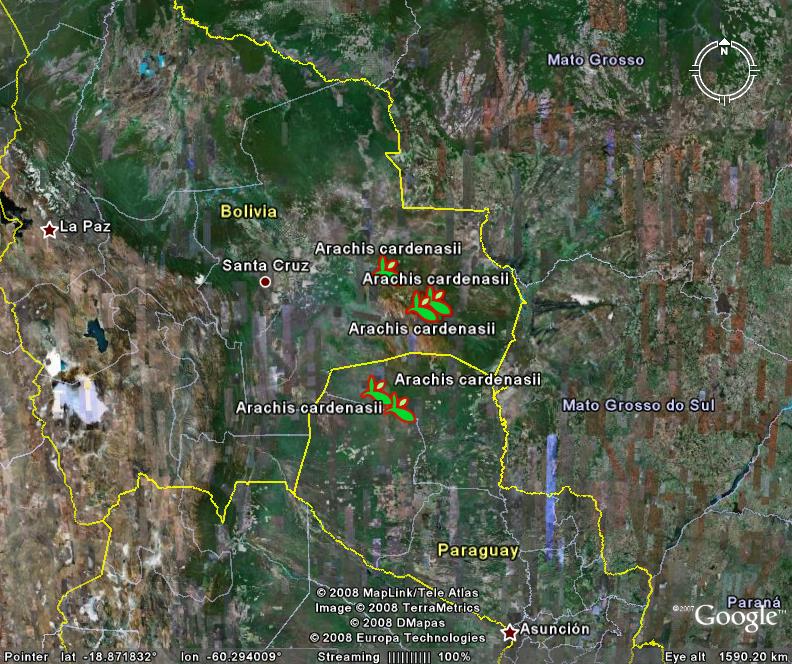
Our friend Andy Jarvis has been explaining to the readers of ICT Update not only how important crop wild relatives are, but also how geographic information systems can help conserve and make use of these important genetic resources. In the context of a longer article on Eco-efficient agriculture, Andy uses the peanut to point out that:
Our friend Andy Jarvis has been explaining to the readers of ICT Update not only how important crop wild relatives are, but also how geographic information systems can help conserve and make use of these important genetic resources. In the context of a longer article on Eco-efficient agriculture, Andy uses the peanut to point out that:
There are, for example, a total of 69 species of crop wild relatives that are in some way related to the cultivated peanut. Of these, 17 species are under significant threat of extinction from the expansion of the agriculture in Brazil, Paraguay, Argentina and Bolivia. Our analyses have demonstrated that a further 15 species are significantly threatened with extinction from climate change.
Jarvis and his team at CIAT use GIS and data from existing specimens to predict where important species might be found.
Collectors can then use global positioning systems (GPS), loaded with the data, to locate the vulnerable species and collect their seed.
Will that be enough to preserve the crop wild relatives on which the future of agriculture depends? Who knows. But it is a start.
P.S. In the same issue, Kwesi Atta-Krah, Deputy Direcctor General of Bioversity International, answers some questions about biodiversity, “the richest natural resource“.