PhyloGenerator is “an open-source, stand-alone Python program, that makes use of pre-existing sequence data and taxonomic information to largely automate the estimation of phylogenies.” Sounds intriguing, no? I found out about it via this Twitter exchange with Rich Grenyer:
@rich_ @Phalaropus Sure, of course!
— AgroBioDiverse (@AgroBioDiverse) July 11, 2013
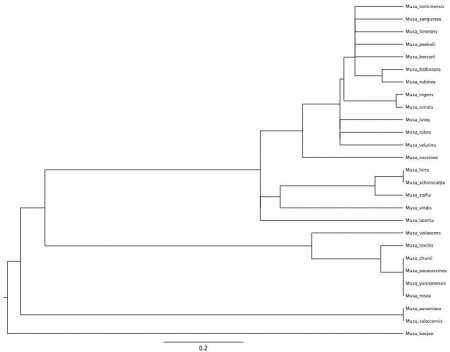
He very kindly shared his automatically generated Musa phylogeny, which is reproduced below. I’m afraid you’ll have to click on the image to read the species names.
So now I’m reaching out to all you banana taxonomists out there. Does this make any sense?