Our friend Danny Hunter at the Crop Wild Relatives Group has word of a new Global Trees Campaign to protect stands of wild trees, among them wild pears and apples in the Caucasus. A project officer has provided Danny with a special report.
Crop maps of Russia and its neighbors
I have often looked for detailed crop distribution maps for the countries of the former Soviet Union and found these hard to come by. Not any more! There is a fabulous on-line atlas of agriculture in Russia and neighboring countries.
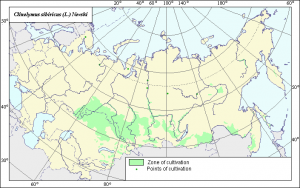
It has descriptions and maps for a 100 crops, including potato and wheat of course, but also lesser known niceties as the Snowball Tree, Sea Bucktorn and Winter Squash. The maps are pretty, here is an example for Siberian Wild Rye (you know, Siberian Black-eyed Susan; Clinelymus sibiricus (L.) Nevski). Better still, they will be available for dowload in GIS format next year.
There are also entries for 540 wild crop relatives and other agriculturally relevant plants, and for pests, diseases and weeds.
Awesome.
Nibbles: Bees, Millennium Villages, Oaks, Wolf, CWR
- “Francis Ratnieks, the UK’s only professor of apiculture, is undertaking pioneering research based on a breed of worker bee genetically programmed to keep hives clean.”
- Scaling up the Millennium Villages. Still no news on what it all means for agrobiodiversity.
- Good news for New England acorn lovers. Including the artisanal pork industry?
- The Ethiopian wolf is in trouble.
- The Crop Wild Relatives Discussion Group reaches 250 member, 200 messages. Well done.
Nibbles: Wild food, Sisal, Cucurbits, Carnival, Rice blight
- Zimbabwean take to wild foods, and not in a good way.
- “It was the afternoon of my eighty-first birthday, and I was in bed with my catamite when Ali announced that the archbishop had come to see my sisal flooring.”
- Gourds+Halloween=Jawdropping Creativity.
- Tangled Bank 117.
- “Terror agent” listing for Xanthomanas oryzae blights US rice research.
Nibbles: Big tent, Certification, Macadamia, Wild harvest, Last meals
- Biodiversity and ecosystems experts to enter big tent: any room for agriculture?
- Japan to recognize sustainable practices that value biological diversity in agriculture, forestry, and fisheries via cute logo.
- Saving drought-resistant macadamia.
- Free food!
- Endangered food.