![]() You may have noticed that a recent Crop Science paper by various ICRISAT staff, entitled “Mini Core Collection as a Resource to Identify New Sources of Variation” ((H.D. Upadhyaya et al. (2013). Mini Core Collection as a Resource to Identify New Sources of Variation Crop Science DOI: 10.2135/cropsci2013.04.0259)) and focusing on chickpea, has been attracting a certain amount of media interest. In brief, Hari Upadhyaya of the ICRISAT genebank and his co-workers evaluated a set of 211 chickpea accessions from the genebank for resistance to a whole slew of stresses, caused by both environmental factors (abiotic) and pests and diseases (biotic), and picked out some real winners. Those 211 chickpeas were not chosen at random. They were carefully selected to reflect the overall diversity of the collection of some twenty thousand samples managed by ICRISAT. Hari explains the rationale for this on the genebank’s chickpea web pages (in fact, he has done similar work on all his mandate crops):
You may have noticed that a recent Crop Science paper by various ICRISAT staff, entitled “Mini Core Collection as a Resource to Identify New Sources of Variation” ((H.D. Upadhyaya et al. (2013). Mini Core Collection as a Resource to Identify New Sources of Variation Crop Science DOI: 10.2135/cropsci2013.04.0259)) and focusing on chickpea, has been attracting a certain amount of media interest. In brief, Hari Upadhyaya of the ICRISAT genebank and his co-workers evaluated a set of 211 chickpea accessions from the genebank for resistance to a whole slew of stresses, caused by both environmental factors (abiotic) and pests and diseases (biotic), and picked out some real winners. Those 211 chickpeas were not chosen at random. They were carefully selected to reflect the overall diversity of the collection of some twenty thousand samples managed by ICRISAT. Hari explains the rationale for this on the genebank’s chickpea web pages (in fact, he has done similar work on all his mandate crops):
The main reason for low use of germplasm in crop improvement programs is the lack of information on large number of accessions, particularly, for traits of economic importance which display a great deal of genotype × environment interaction. Brown (1989) introduced the concept of developing core collection, which consist of about 10% of entire collection and represent at least 70% of the genetic variability of the entire collection, as a gateway to the enhanced utilization of germplasm in breeding. Core collection in chickpea has been reported (Upadhyaya et al. 2001). However, it soon became evident that developing core collections will not solve the problem of low use of germplasm, as even the size of core collection could still be large. To overcome this, Upadhyaya and Ortiz (2001) proposed mini core collection concept (consisting of 10% of the core or 1% of the entire germplasm), and developed mini core subsets in chickpea (Upadhyaya and Ortiz 2001).
The extensive evaluation of the chickpea mini core collection described by Hari and his colleagues in the paper, which spanned many sites and several years, allowed them to identify 40 accessions which were resistant to abiotic stresses, 31 to biotic stresses and 6 accessions which were in both groups. Very useful information for any chickpea breeder, who can now request material by name (or rather, accession number) rather than sending vague requests to Hari for material with resistance to this or that. This sort of work by genebanks makes life easier for breeders and researchers, and increases use of the collection. Which everyone wants to see, not least the money people.
However, even if you get hold of a PDF of the original paper, you may still have a few questions. And so we rolled up our sleeves on your behalf and made the necessary journey into Genebank Database Hell.
One question you may have is: what accessions are in the chickpea mini core? The paper doesn’t list all 211, but you can get the full list from one of the ICRISAT genebank’s web pages. Just click on the link on that page and you’ll see accession number and country of origin for all 211 accessions.
The obvious next question is: how do I get more information on all those accessions? After all, you may want to do your own analysis of the evaluation results. Well, you could cut and past those accession numbers into ICRISAT’s online genebank database. But as chance would have it, there is an easier way, as USDA has a safety duplicate of the mini core, and a set of ICRISAT’s evaluation results is also online at GRIN. It might not be as bang up to date as what’s in the paper, but it’s a start.
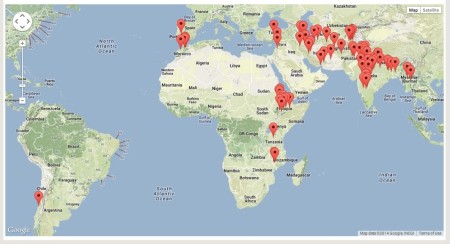
Next: what’s the geographic distribution of the accessions in the mini core. Ok, so for this, you could just have a look at the countries of origin, but really dots on a map would be best, right? That turns out not to be so easy to get, you may or may not be surprised to find. To cut a long story short, you will be able to do it in the next iteration of Genesys, but that’s not public yet. Fortunately, I was able to sneak into the sandbox, and for the 25% or so of the 211 accessions which actually have georeferences, this is the result. You saw it here first, folks.
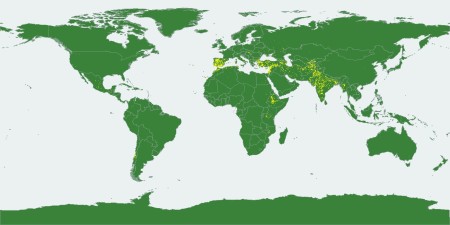
Compare to the distribution of the third of all 48,000-odd chickpea accessions listed in Genesys (current version) which can be mapped.
Yes, I know it’s a little difficult to compare, but that’s really the best I can do at the moment. One question you may have based on that is to what extent the mini core is representative of the climates from which chickpeas have been collected. But that I really cannot answer at the moment without devoting some serious effort to it.
One last thing, which is a little bit more esoteric, so let me sound a Genebank Wonk Alert. You have been warned. Delve a little deeper into the data on one of those 6 super-chickpeas with a whole bunch of resistances, ICC 14402, and you find that, unlike the other 210, which are all labelled as landraces, this one is described as “Breeding/Research material.” What is its history? It’s a little tricky to piece together, but if you look at the other accession numbers which are associated with that accession, both in the ICRISAT database and on GRIN, you get the impression that it represents a selection from a Turkish landrace, ICC 4490. It’s impossible to prove that from the data available online, but I verified that impression with Hari, and he confirmed it in an email:
…ICC 4490 (P 5924; 88 1-4 Spanish) is a landrace from Turkey and was donated in 1973 to ICRISAT. At ICRISAT this line (ICC 4490) was screened in wilt sick nursery and a wilt resistant single plant was selected from this line. The original accessions was maintained as ICC 4490 and new accession identity ICC 14402 was given to this wilt resistant selection in 1986. WR stand for “Wilt resistant”.
There’s clearly quite a lot on information that hasn’t made the journey from the brains of genebank curators to the relevant databases, or indeed the published literature. That’s a little worrying to me.
The idea of ‘core collections’ was suggested by Frankel with a specific purpose in mind: to provide, by means of careful sampling, a subset of manageable size from a large germplasm collection, for testing General Combining Ability (GCA) [Frankel, O.H. 1986 Genetic conservation in perspective. In. V.L. Chopra & T.N. Khoshoo (eds.) Conservation for Productive Agriculture. ICAR, New Delhi, pp. 50-57.]. Crosses were not feasible on tens of thousands of accessions:
Without using the term ‘core’, subsets of different sizes representing the variability in the collection had been used at IITA for cowpea accessions in the late 1970s. From this very rational origin, the concept of cores has rapidly changed its nature under strong promotion, and now threatens the integrity of carefully assembled germplasm collections. ‘New’ cores attempt to be sets of samples adequately representing variation for genetic and evaluation studies [Brown, A.H.D. 1989 Core collections: a practical approach to genetic resource management. Genome 31:818-824.] representation of wild species: the perennial Glycine collection at CSIRO is a typical example. The concept of crossability — the purpose of Frankel’s concept of cores — has been entirely ignored, as wild relatives from secondary and tertiary gene pools may be included (for example, the European Cooperative Programme barley core collection will include 50-80 accessions of wild species for research purposes).