Our friend, colleague and occasional contributor Andy Jarvis received GIBF‘s Ebbe Nielsen Prize for innovative bioinformatics research last night in Copenhagen. Andy has been doing his trademark work on the spatial analysis of crop wild relative distributions ((The maps below are an example. They show the modeled distribution of species richness in the genus Phaseolus now, and then in 2050.)) at CIAT, just outside Cali in Colombia, jointly with Bioversity International. He used the occasion to highlight the contribution made to this effort by his numerous Colombian colleagues. Congratulations to all of them.
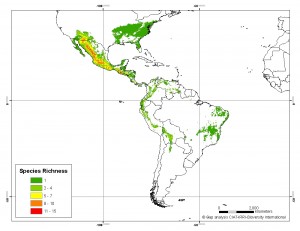
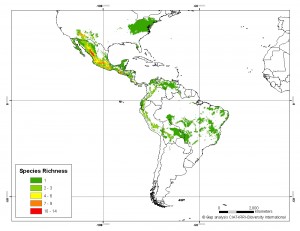
LATER: Here’s Andy’s talk.
The gap analysis presented for Zea is rather odd. Comments left on the web page seem to go unnoticed and unposted, so at the risk of double-posting, I copy the comments here:
I’d also like to point out some issues with the Zea analysis. It’s not clear to me whether USDA GRIN was checked for germplasm (though #18 on the list would have me believe it was), because my interpretation of the results in GRIN lead me to nearly opposite conclusions: 226 parviglumis accessions available (52 active and available for ordering), but only 13 Zea perennis accessions (4 active) and 26 Zea luxurians accessions (only 2 active!).
Moreover, some of the distributions are surprising — Zea diploperennis, for example, seems to have been collected in several parts of Mexico, and gap analysis shows it’s potential distribution throughout Jalisco. But to my knowledge, Zea diploperennis is only known from the Sierra de Manantlan Biosphere reserve. The potential distributions of other wild taxa also seem off. In fact, we have a pretty good record of collections of most of these taxa — I urge those interested to read Ruiz et al (2001), Ruiz et al. (2008), and Sanchez-Gonzalez & Ruiz Corral (1997), the latter of which is publicly available online from CIMMYT:
http://www.cimmyt.org/english/docs/proceedings/geneflow/Geneflow_TeosDistr.pdf
Ruiz JA, Sanchez JDJ, Aguilar M (2001) Potential geographical distribution of teosinte in Mexico: A GIS approach. Maydica 46: 105-110.
Ruiz Corral JA, Duran Puga N, Sanzhez Gonzalez JD, Ron Parra J, Gonzalez Eguiarte DR et al. (2008) Climatic adaptation and ecological descriptors of 42 Mexican maize races. Crop Sci 48: 1502-1512.
Sánchez González JJ, Ruiz Corral JA (1997) Teosinte distribution in Mexico. In: Serratos JA, Willcox MC, Castillo González F, editors. Gene flow among maize landraces, improved maize varieties and teosinte: implications for transgenic maize. Mexico City: CIMMYT. pp. 18-36.
Hi Jeff,
Many thanks for your pretty interesting and useful feedback. It’s actually what we seek through the portal. I have not been noticed by my webmaster whether there’s a comment which is still unposted, and I do not see any comment unattended. I will check deeper though, as I wouldn’t like to lose these important feedbacks.
One of the issues with our approach is that we strongly depend on freely available data from databases (e.g. GRIN, SINGER, EURISCO, GBIF). We thus acknowledge there should be hidden accessions that might change the priorities picture. Moreover, we also depend on the quality of such information and definitely on the presence of location data (e.g. lat/lon, or locality and country names) within the accessions/specimens. In our approach, samples with no georreferences cannot be assessed in any way as they do not provide any useful information (neither geographical, nor environmental, or genetic, or agronomic) more than a taxon name. So, we just discard them in one of the first steps. Perhaps that’s where most of the Z. parviglumis accessiones went?
Moreover, the method also takes into account the geographical and environmental sampling/collection bias versus the potential area in which the taxon is distributed. So, if the taxon is potentially distributed within a considerably big area, no matter if you have 5,000 accessions, but if they have been collected at only one site (or within an small part of the potential area) then your taxon will be flagged as underrepresented.
Some of the data is still hidden, and you point out additional resources which we will query in order to re-assess the Zea gap analyses and correct the results. We know there’s a lot of work to be done yet, and we continue working and improving our results. This information you give us is actually what we need to be more accurate. Many thanks again for your comment
@Julian
I just chanced across the gapanalysis site and had a look at the crops that I am familiar with, leaving comments for Hordeum and Vicia [probably better described as Vicia faba et al.].
With only 10-20 % of passport data having georeferences, the conclusions which can be drawn from gapanalyses with regard to conservation status are rather limited at the moment.
Many thanks for your contributions, they are pretty useful for us. We are seeking for data in order to improve our results. But in some cases you just need to conclude there’s not enough ecogeographic knowledge on a genepool or set of taxa to perform such an analysis. It’s quite frustrating in some cases to be unable to get out of genebank database hell.
There are some genepools for which data is still limited, and in those cases, conclusions are also limited, but for other genepools you just find out that conclusions are interesting in view of ex-situ conservation planning. And it’s quite exciting to speak with an expert and verify that your results and conclusions are accurate.
Please take a look to this recent presentation we prepared for Cary Fowler head of the Trust. It contains current results for two sections of the Phaseolus.
Slide 11: where is the complementarity between in-situ and ex-situ conservation?
Hi again,
Thanks for the link to slide share. It made my day.
There are a fair few data for barley in the European barley database. Check out the final presentation from the EU GENRES 104 project on utilisation of barley genetic resources: Barley genetic resources
Looks like you’ve done a pretty huge work on Barley genetic resources. It’s a pretty interesting presentation. Are all those characterisation data available to fully download?
There is no download function, not all the data used for presentation are in the database.
There is certainly scope for collaboration if anyone is interested.
What are you thinking of?
Definitely. We’re currently engaged in this project and aim to evaluate the completeness of genebank holdings. We seek for passport as well as characterisation data on cropped accessions of some species, including barley. However, all we can offer is publication co-authorship as our budget is already defined. If you wish to work with us it would be great, as this initial work might lead to further refining of methods, validation of results, which could be justified within a new project phase.