Future PGRFA collections will focus on filling gaps in existing collections, collection of certain regional, minor and subsistence crops and collection from particular countries where collection has not taken place or been very limited.
That’s from the Global Plan of Action for the Conservation and Sustainable Utilization of Plant Genetic Resources for Food and Agriculture (GPA). In fact, “gap-filling” is often mentioned as a way to be more strategic and cost-efficient in germplasm collecting. This approach relies on knowing where a crop (or, more correctly, landraces of a crop) is grown, and comparing that with the distribution of germplasm accessions in genebanks (which could in fact be done in various different ways, depending on how you define “gap”, but forget that for a minute).
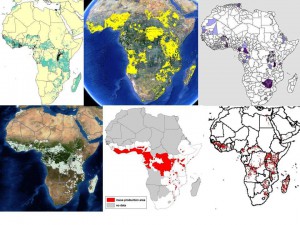
We all know about the problems associated with data on germplasm accessions (lack or inaccuracy of georeferences in passport data, for example). But in fact there are issues with the crop distribution data too, as we’ve recently discussed here and here. This shows the range of answers you get when you ask the seemingly simple question: where do bananas grow in Africa? Click on the image to see it better, or go to the previous posts.
So, is “gap-filling” a forlorn hope, at least at the continental and global levels? Looking forward to your thoughts…
I guess that depends on how much resources (i.e. time, money) you want to invest in getting the data in the best shape for gap-filling analyses, and also on how many crops, which crops. To illustrate my point:
For potato, you have at least three crop distribution surfaces developed with different methods (as you have for bananas), you have FAO data and possibly national level statistics, plus you have Hijmans (2001) crop distribution data. Yet you have to figure out which source is best, combine all of them, and probably fix some errors in the distribution surface, or update the data. You have access to landrace data from the CIP genebank which is very likely to have reliable coordinates, at least for the Andes. You invest a bit of time on basic data cleaning and organisation, and then apply the method. Then you get the gaps for the Americas: done!.
For finger millet, you do not have any crop distribution surface, and most of the data in FAO remains aggregated as “millets”. Besides that, collections are somehow limited and data from major collections (Asian research centres) remains hidden. Yes, you have the CGIAR collections, but these might only be complementary to other major or national collections, particularly in India and Sub-Saharan Africa. So, how much time and other resources are required to: (1) get genebank data out, clean it, and correct it if necessary, (2) know where the crop is grown?
But I’m still trying to get a potato-type example but at the global level. Any ideas?
The GBIF distributed data network could provide useful occurrence data for the crop species from many other sources than the genebank collections. Even if these occurrence data points unfortunately very often have uncertainties and other issues of data quality, they could give additional information to guide the identification of gaps in the genebank collections. At the TDWG 2009 conference Jarvis et al (2010) presented their experiences from modeling crop species from the GBIF “data-pool”, http://www.tdwg.org/proceedings/article/view/555. I think this would be a fruitful path to explore further. National biodiversity survey data shared with GBIF (from data publishers outside the genebank community) could probably also help to complete the picture of where a crop is currently grown…
Surely there won’t be much in the way of crop data in GBIF? Crop wild relatives, sure. But cultivated material? I doubt there’s anything there besides the genebanks.
I believe that we will find also some stray samples from crop species in herbaria and history collections (as Dirk also mentions below for Vicia). I made a brief look at barley and wheat and did find some samples of Hordeum vulgare and Triticum aestivum in non-genebank collections in Sweden and Norway. There seems to remain major issues with application/reporting of the proper naming (nomenclature) of the reported species to navigate through though…!
If nothing else – it might perhaps be easier to encourage non-genebank natural collections with stray samples of landraces and other crop plants to share them with GBIF – than to share them to a dedicated PGR information system…?
Very many small streams (stray samples from a large number of collections) could perhaps make enough interesting crop data points to make it worthwhile to gather them and to have a look?
Yes, you’re right. The more sources you have into GBIF, the better it gets for these type of analyses. The problem is that most of the collections are still hidden even to GBIF. In addition: taxonomies, coordinate accuracy, sample identification, among others, are issues that need to be tackled whenever possible. Sometimes institutions intentionally “degrade” their data by removing decimal places in the coordinates, or by removing the coordinate as a whole. In particular, herbarium specimens and observations of landraces can aid with crop-areas identification, however, the issue of when the sample was recorded remains to be important. I guess you’d need to work out in a case-by-case basis and see what you can get.
This again has something to do with genebank database hell, we need to work out the data gap first.
I think you are barking up the wrong tree. A rough knowledge of where a crop grows should be enough. Diversity of a crop is not strongly related to global crop density. The bottleneck is to understand (have a plausible model) of the distribution of diversity in the crop, and sample from that.
But doesn’t a plausible model for the geographical distribution of diversity in a crop sort of depend on a plausible model for the geographical distribution of the crop?
Most crop distribution datasets rely in the last instance on agricultural censuses, which have more than one problem. Rare crops are usually underreported, for instance. And as Julian indicates, different species may be lumped as one crop.
Crop density is not necessary to have (Robert), but we need rather precise data on the presence/absence of crops to target collection (Luigi). On the other hand, we also need data on whether varieties are modern or traditional to know if they are relevant for ex situ collections. Traditional varieties are important to have, (recycled) modern varieties are generally not.
Since both distinctions are binary (presence/absence, modern/traditional) I think this is a great case in which on-line crowdsourcing (à la http://www.oldweather.org) could be very powerful. People can then indicate on an on-line map for each grid cell if a crop is present or not, and if (at least some of) the varieties there are traditional.
The questions are simple enough to be answered en masse. I would guess that the result will be more complete than most census data.
But again, I think the problem is the will of data sharing. Genebanks and herbaria do not share their data, or don’t spend any time in organizing, digitizing, georreferencing, etc., and they also tend to hide part (or all) of their data. But let’s suppose you have the data, what you need is:
1. A method
2. A crop landrace presence absence/absence surface
3. a set of genebank crop landrace accessions
Methodological basis could be: (a) the gap analysis website, Endresen (2010), Van Etten & Hijmans (2010), and Ramirez-Villegas et al (2010), among others.
For crop landrace presence/absence surface, a good start point could be the datasets that already exist, plus a quick survey to experts, or perhaps looking into the dates of genebank/herbarium collections can give a clue of whether a particular land area is likely to be sown (partly) with landraces? the newer the accession the more likely the place to still hold landraces (but, can you extra or interpolate this?)
And for the accessions themselves, you all know the answer, probably.
Dagterje is right about GBIF. The data are very useful for modelling species distributions. I recently looked at their distribution data for Vicia species and found some areas which look very promising to fill gaps in current ex situ collections, so, for minor crops GBIF data are handy.
There are probably more passport data available in herbaria that could be linked with seed accessions ie. VIR and other institutes that have a herbarium specimen for each seed accession, in order to fill the gaps in documentation, however this requires resources.
Thank you Luigi for kicking this discussion of. It is indeed an important matter and its value will indeed largely dependent on how you define a “gap”! First of all it is clearly done from an ex situ perspective. The “gap” is equated to material/diversity that is NOT in the collections but available in nature or on farms. I would go along with this perspective.
Regarding the definition, is gap defined from a geographical perspective “only” or are also botanical (i.e. representation of the various species that make up the genepool?) and genetic (i.e. the representation of varieties/genotypes)? I guess it will be both! Furthermore, in using the term gap you imply that you know “what is out there”, but do we really know?
In conclusion, I would argue that we have to take the gap-filling approach with a pinch of salt as in many instances our knowledge of the diversity that makes up the genepool and its distribution is very limited.
Jan
About “you imply that you know “what is out there”, but do we really know?”
There are generally fairly strong geographic patterns in the distribution of genetic diversity (intraspecies). We are working on models to predict diversity, even for unsampled places.
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0012060
And as per your definition of gap: yes, I think a gap needs to be assessed in different dimensions:
(a) species into a genepool and races into a domesticated species,
(b) geographic range (and collected range),
(c) abiotic traits,
(d) specific characteristics that are more desirable (e.g. more gluten content for wheat)
(e) genetic variability within a trait (i.e. which genes do and how do they drive the expression of a certain characteristic)?
Basic ecogeographic gap analysis of crop genepools can be done for a list of taxa, as done here. But the deeper you go, the more data you need.
Jan Engels just put his finger in the wound when he says that “in many instances our knowledge of the diversity that makes up the genepool and its distribution is very limited”.
I would also add that our knowledge of what we maintain and its characteristics is also somehow limited. That is due to the many problems that, chronically, afflicts genebank activities, namely lack of capacity building, human and financial resources, etc., etc. that also jeopardises the sustainable conservation of the material itself.
Time flies and its urgent to complete germplasm collections through gap filling (geographical, botanical and genetic) as a manner of maximising the already very scarce resources before it’s too late, but we shouldn’t also forget to secure the conservation of the material that we already have as well as to deepen the understanding of the diversity already captured. These two premises are paramount to identify the collections’ gaps and the need to filling it by collecting what, from where and when!
I partly disagree with you.
Characterization is very important, of course to make use of the germplasm. However, I think that for gap analysis we don´t have to know everything.
Actually, there is a much more fundamental uncertainty: we don´t know what kinds of traits are needed in the future, especially since diseases evolve over time. Also, we will never know what is in store for us. Traits need to be discovered.
However, with some biological principles and knowledge about the genesis of crop biodiversity, we might well be able to optimize ex situ collection strategies and capture as much biodiversity as possible given limited resources, without knowing exactly what that entails in terms of traits.
I think you’re partially right!
“Characterization is very important, of course to make use of the germplasm. However, I think that for gap analysis we don´t have to know everything.” it’s true when we talking about geographical and botanical gap filling, but when we’re talking about genetic gap filling I think it matters
Genetic gap-filling: molecular diversity can be modelled fairly well with “neutral” models of evolution, though. So we need to know very little…
So, you don’t need to characterize genetically all your collection before going to the field to collect, right?
No, not the whole collection, just enough to be able to develop a model of the geographical genesis of crop diversity. You then select your sampling sites based on the predictions of that model for places that have not been sampled yet. You may come across a few surprises (more or less diversity than expected), but at least you work with plausible expectations.
Agreeing with Robert way up above, its likely that for most crops (excepting perhaps some of the most ‘underutilized’) we have a good idea already where the overwhelming majority of the genetic diversity lies- in the primary region(s) of diversity where the crop has been around for a very long time. So, if we are aiming for efficiently and effectively collecting the widest genetic diversity possible of the crop, it doesn’t seem that terribly complicated to not get too caught up in the details of what, on the genetic level, is already in collections, but rather to make sure that collections have a very good geographic coverage of the whole primary region(s) of diversity. If this region is very large or expensive to collect, then it probably makes increasing sense to research the genetic diversity of what is already in collections and make more targeted collecting efforts.
What gets more complicated, and makes crops very different from most wild species, is that they have been moved around by people and so after some time secondary regions of diversity have developed, and perhaps these regions may hold particular diversity for paticular traits of interest? And how good is our knowledge of this diversity?
Thanks for this very interesting conversation.
I guess that works in a first phase of geographic gaps filling, but how would you do it when you have all the geographic distribution of the crop already sampled? you could end up just sampling more where your model predicts more diversity, or just not knowing what to sample when you get to the collection site.
So, I agree with Colin in that in a first collection effort, the genetic level (though it can certainly add value to the collecting efforts and better target or focus them) might not be that necessary.
True, for smaller collections some basic geographic gap filling might be a good first step. To ensure coverage up to some magic percentage (“conserving 70% of the current genetic diversity of crop X”) for the big crops, we would need some genetic work, though.
Colin, I don´t think this is what Robert writes above?
Geographic coverage isn´t everything. For geographic representativeness, even-spaced sampling across the range of each crop would be enough. However, the collection is more diverse if it has more sampling density in regions where genetic diversity is highest. Current collections might not always be balanced that way and we would need to put numbers on diversity to really know what is going on.
Diversity is generally highest around centres of origin but “surfing” processes during crop range expansion and selection by humans can create “neodiversity” away from the centre of origin. Perhaps we have way proportionally too much diversity from centres of origin and should start sampling more outside of the centres? Who knows.
As commented above the genetic diversity is not distributed even across spatial space, but probably concentrated in the center of origin (often renamed as center of diversity based on the interpretation of Vavilovs intentions with this concept). So using the ecological niche modeling approach to evaluate if the genebank collections are well represented across the ecological space, could thus make sense with the aim to capture genetic diversity with the adaption to different ecogeographies.
* Note that this approach is not the same as sampling across the variation of genetic diversity because certain ecoregions might be have more genetic diversity…
* And that adaption to specific ecogeography – different from the ecogeography of the center of origin/diversity might sometimes be economically valuable/useful. Eg for cultivation in new, unusual or more marginal areas…?
* And what if the FIGS approach can help us to identify ecoregions for such a target trait property. Perhaps a crop improvement program need a particular trait property. Perhaps this trait property is not typical of the genetic diversity at the center of origin/diversity. Perhaps a FIGS analysis could assist your germplasm collecting expedition to the locations where this trait property is more likely to be found…?
Just a thought.